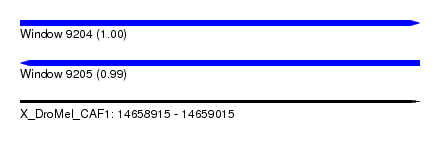
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,658,915 – 14,659,015 |
| Length | 100 |
| Max. P | 0.996379 |

| Location | 14,658,915 – 14,659,015 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -30.42 |
| Energy contribution | -30.42 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14658915 100 + 22224390 GAGAGCAAGCGAGUUGUAAUUGAAUGUUUGGCUCUUAAUUAAAACAAGAAUUGUGACAGCCACGCUUUAAGGCGG--GGGGCGUGGCAAGCCCAAGCCUAAG .(((((..((..((..(((((...(((((............)))))..)))))..)).))...))))).((((..--.((((.......))))..))))... ( -29.30) >DroSec_CAF1 123092 94 + 1 GAGGGCAAGCGAGUUGUAAUUGAAUGUUUGGCUCUUAAUUAAAACAAGAAUUGUGGCAGCCACGCUUUAAGGCGG--GGGGCGUGGCAAGCCCAAG------ ..((((......((..(((((...(((((............)))))..)))))..)).((((((((((.......--))))))))))..))))...------ ( -33.60) >DroEre_CAF1 116363 93 + 1 GAGGGCAAGCGAGUUGUAAUUGAAUGUUUGGCUCUUAAUUAAAACAAGAAUUGUGGCAGCCACGCUUUAAGGCG---GGGGCGUGGCAAGCCCGGU------ ..((((......((..(((((...(((((............)))))..)))))..)).((((((((((......---))))))))))..))))...------ ( -34.10) >DroYak_CAF1 118994 93 + 1 GAGGGCAAGCGAGUUGUAAUUGAAUGUUUGGCUCUUAAUUAAAACAAGAAUUGUGACAGCCACGCUUUAAGGCGGAGGGGGCGUGGCAAGCCC--------- ..((((......((..(((((...(((((............)))))..)))))..)).((((((((((.........))))))))))..))))--------- ( -31.10) >consensus GAGGGCAAGCGAGUUGUAAUUGAAUGUUUGGCUCUUAAUUAAAACAAGAAUUGUGACAGCCACGCUUUAAGGCGG__GGGGCGUGGCAAGCCCAAG______ ..((((......((..(((((...(((((............)))))..)))))..)).((((((((((.........))))))))))..))))......... (-30.42 = -30.42 + 0.00)


| Location | 14,658,915 – 14,659,015 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -22.95 |
| Consensus MFE | -21.87 |
| Energy contribution | -21.68 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14658915 100 - 22224390 CUUAGGCUUGGGCUUGCCACGCCCC--CCGCCUUAAAGCGUGGCUGUCACAAUUCUUGUUUUAAUUAAGAGCCAAACAUUCAAUUACAACUCGCUUGCUCUC ....(((((((((..(((((((...--..........))))))).))).))).(((((.......))))))))............................. ( -24.02) >DroSec_CAF1 123092 94 - 1 ------CUUGGGCUUGCCACGCCCC--CCGCCUUAAAGCGUGGCUGCCACAAUUCUUGUUUUAAUUAAGAGCCAAACAUUCAAUUACAACUCGCUUGCCCUC ------...((((..(((((((...--..........)))))))........((((((.......)))))).........................)))).. ( -22.52) >DroEre_CAF1 116363 93 - 1 ------ACCGGGCUUGCCACGCCCC---CGCCUUAAAGCGUGGCUGCCACAAUUCUUGUUUUAAUUAAGAGCCAAACAUUCAAUUACAACUCGCUUGCCCUC ------...((((..(((((((...---.........)))))))........((((((.......)))))).........................)))).. ( -22.90) >DroYak_CAF1 118994 93 - 1 ---------GGGCUUGCCACGCCCCCUCCGCCUUAAAGCGUGGCUGUCACAAUUCUUGUUUUAAUUAAGAGCCAAACAUUCAAUUACAACUCGCUUGCCCUC ---------((((..(((((((...............)))))))........((((((.......)))))).........................)))).. ( -22.36) >consensus ______CUUGGGCUUGCCACGCCCC__CCGCCUUAAAGCGUGGCUGCCACAAUUCUUGUUUUAAUUAAGAGCCAAACAUUCAAUUACAACUCGCUUGCCCUC .........((((..(((((((...............)))))))........((((((.......)))))).........................)))).. (-21.87 = -21.68 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:36 2006