
| Sequence ID | X_DroMel_CAF1 |
|---|---|
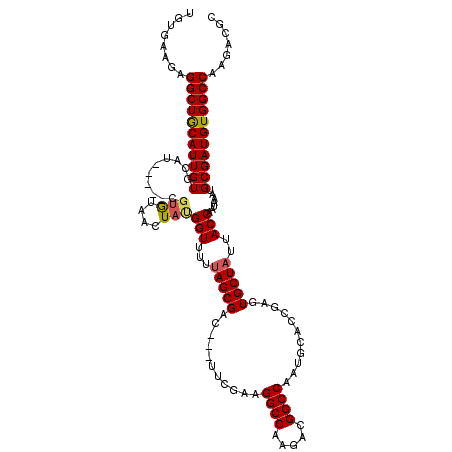
| Location | 14,652,451 – 14,652,603 |
| Length | 152 |
| Max. P | 0.945780 |

| Location | 14,652,451 – 14,652,563 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -35.49 |
| Consensus MFE | -29.99 |
| Energy contribution | -30.15 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
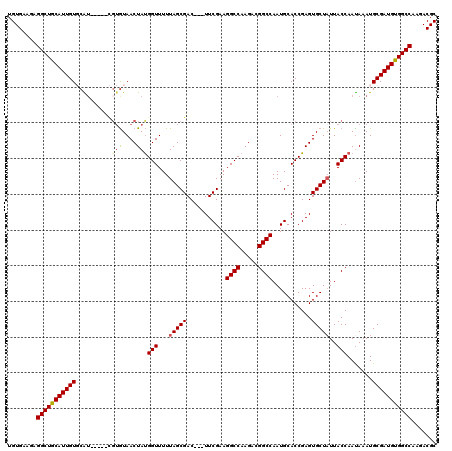
>X_DroMel_CAF1 14652451 112 + 22224390 UGUGAAGAGGCUGCAUUGUGCAG-----CGUGUAACUAUGGUUUUUAGCGAC---UUCGAAGGCCAAGACGGCCAAUGCACCGAGUGCUCUUACCAAUAAAUGCGAUGUGGCCAAGACGC ........(((..((((((((((-----(....(((....)))....))...---......((((.....))))..))))).....((..(((....)))..))))))..)))....... ( -32.00) >DroSec_CAF1 121764 112 + 1 UGUGAAGAGGCUGCAUUGUGCAU-----CGUAUAACUACGGUUUUUAGCGAC---UUCGAAGGCCAAGACGGCCAAUGCACCGAGUGCUAUUACCAAUAAAUGCGAUGUGGCCAAGACGC ........(((..((((((....-----.(((....)))(((...((((.((---((((..((((.....))))..))....))))))))..))).......))))))..)))....... ( -34.50) >DroSim_CAF1 120509 112 + 1 UGUGAAGAGGCUGCAUUGUGCAU-----CGUGUAACUAUGGUUUUUAGCGAC---UUCGAAGGCCAAGUCGGCCAAUGCACCGAGUGCUAUUACCAAUAAAUGCGAUGUGGCCAAGACGC ........(((..((((((((..-----...)).....((((...((((.((---((((..((((.....))))..))....))))))))..))))......))))))..)))....... ( -34.10) >DroEre_CAF1 115125 112 + 1 UGUGAAGAGGCUACAUUGUUCGU--------GUAACUAUGGUUUUUAGCGACCAAUUCGAAGGCCAAGACGGCCAAUGCACCGAGUGCUAUUACCAACAAAUGCGAUGUGGCCUAGACGC .(((...((((((((((((...(--------((.....((((...(((((...........((((.....))))...........)))))..)))))))...))))))))))))...))) ( -38.95) >DroYak_CAF1 117806 117 + 1 UGUGAAGAGGCUGCAUUGUUCGUGUGUGUGUGUAACUAUGGUUUUUAGCGAC---UUCGAAGGCCAAGAUGGCCAAUGCACCGAGUGCUAUUACCAAUAAAUGCGAUGUGGCCUAGACGC .(((...((((..((((((...(((..((.....))..((((...((((.((---((((..((((.....))))..))....))))))))..)))))))...))))))..))))...))) ( -37.90) >consensus UGUGAAGAGGCUGCAUUGUGCAU_____CGUGUAACUAUGGUUUUUAGCGAC___UUCGAAGGCCAAGACGGCCAAUGCACCGAGUGCUAUUACCAAUAAAUGCGAUGUGGCCAAGACGC ........(((((((((((..........(((....)))(((...(((((...........((((.....))))...........)))))..))).......)))))))))))....... (-29.99 = -30.15 + 0.16)

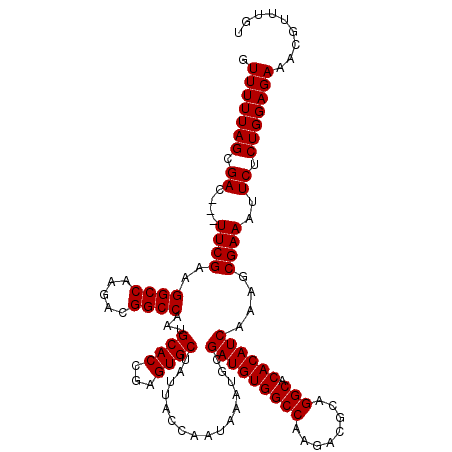
| Location | 14,652,486 – 14,652,603 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.84 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -33.00 |
| Energy contribution | -33.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
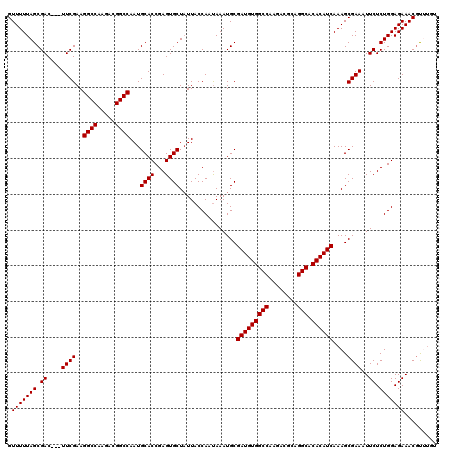
>X_DroMel_CAF1 14652486 117 + 22224390 GUUUUUAGCGAC---UUCGAAGGCCAAGACGGCCAAUGCACCGAGUGCUCUUACCAAUAAAUGCGAUGUGGCCAAGACGCAGGCACACAUCAAAGCGAAAUUCUCUGGAGAAACGUUUGU .(((((((.((.---((((..((((.....))))...((((...))))................(((((((((........))).))))))....))))..)).)))))))......... ( -33.00) >DroSec_CAF1 121799 117 + 1 GUUUUUAGCGAC---UUCGAAGGCCAAGACGGCCAAUGCACCGAGUGCUAUUACCAAUAAAUGCGAUGUGGCCAAGACGCAGGCACACAUCAAAGCGAAAUUCUCUGGAGAAACGUUGGU .....((((.((---((((..((((.....))))..))....))))))))..((((((...((((((((((((........))).))))))...)))...((((....))))..)))))) ( -33.50) >DroSim_CAF1 120544 117 + 1 GUUUUUAGCGAC---UUCGAAGGCCAAGUCGGCCAAUGCACCGAGUGCUAUUACCAAUAAAUGCGAUGUGGCCAAGACGCAGGCACACAUCAAAGCGAAAUUCUCUGGAGAAACGUUUGU .(((((((.((.---((((..((((.....))))...((((...))))................(((((((((........))).))))))....))))..)).)))))))......... ( -33.00) >DroEre_CAF1 115157 120 + 1 GUUUUUAGCGACCAAUUCGAAGGCCAAGACGGCCAAUGCACCGAGUGCUAUUACCAACAAAUGCGAUGUGGCCUAGACGCAGGCACACAUCAAAGCGAAAUUCUCUGGAGAAACGCAUCU .......(((.....((((..((((.....))))...((((...))))................((((((((((......)))).))))))....)))).((((....)))).))).... ( -35.00) >DroYak_CAF1 117846 117 + 1 GUUUUUAGCGAC---UUCGAAGGCCAAGAUGGCCAAUGCACCGAGUGCUAUUACCAAUAAAUGCGAUGUGGCCUAGACGCAGGCACACAUCAAAGCGAAAUUCUCUGGAGAAACCCAUUU .(((((((.((.---((((..((((.....))))...((((...))))................((((((((((......)))).))))))....))))..)).)))))))......... ( -34.40) >consensus GUUUUUAGCGAC___UUCGAAGGCCAAGACGGCCAAUGCACCGAGUGCUAUUACCAAUAAAUGCGAUGUGGCCAAGACGCAGGCACACAUCAAAGCGAAAUUCUCUGGAGAAACGUUUGU .(((((((.((....((((..((((.....))))...((((...))))................(((((((((........))).))))))....))))..)).)))))))......... (-33.00 = -33.00 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:34 2006