
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,639,197 – 14,639,295 |
| Length | 98 |
| Max. P | 0.966079 |

| Location | 14,639,197 – 14,639,295 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.62 |
| Mean single sequence MFE | -23.55 |
| Consensus MFE | -14.70 |
| Energy contribution | -13.93 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
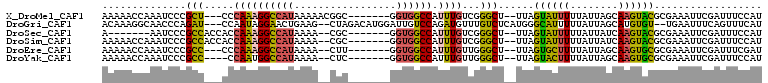
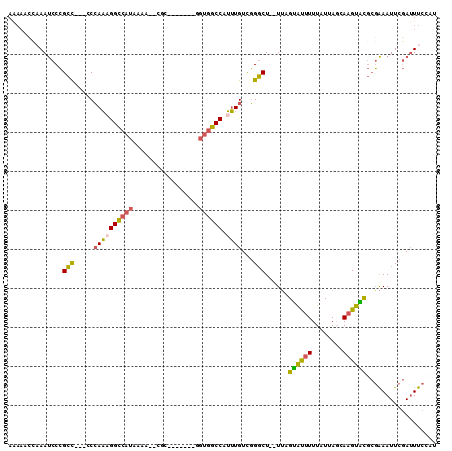
>X_DroMel_CAF1 14639197 98 + 22224390 AAAAACCAAAUCCCGCU---CCCAAAGGCCAUAAAAACGGC-------GGUGGCCAUUUGUCGGGCU--UUAGUAUUUUUAUUAGCAAGUACGCGAAAUUCGAUUUCCAU .......(((((..(((---(.((((((((((.........-------.)))))).))))..)))).--...((((((........)))))).........))))).... ( -20.90) >DroGri_CAF1 117577 103 + 1 ACAAAGGCAACCCAGAU---CCAAUAGGAACUGAAG--CUAGACAUGGAUUGUCCAGAUGUUUGUCUCAUGGGCAUUUUUAUUAGCAUGUGU--UGAAUUUCAGUUUCAU ..(((..((((.(((.(---((....))).)))..(--((((....(((.((((((((........)).)))))).)))..)))))....))--))..)))......... ( -22.80) >DroSec_CAF1 109003 92 + 1 A-------AAUCCCGCCACCACCAAAGGCCAUAAAA--CGC-------GGUGGCCAUUUGUCGGGCU--UUAGUAUUUUUAUUAUCAAGUACGCGAAAUUCGAUUUCCAU .-------.....(((..((..((((((((((....--...-------.)))))).))))..))...--...((((((........)))))))))............... ( -21.80) >DroSim_CAF1 106756 99 + 1 AAAAACCAAAUCCCGCCACCACCAAAGGCCAUAAAA--CGC-------GGUGGCCAUUUGUCGGGCU--UUAGUAUUUUUAUUAUCAAGUACGCGAAAUUCGAUUUCCAU .............(((..((..((((((((((....--...-------.)))))).))))..))...--...((((((........)))))))))............... ( -21.80) >DroEre_CAF1 102712 96 + 1 AAAAACCAAAUCCCGCC---CCCAAAGGCCAUAAAA--CUU-------GGUGGCCAUUUGUUGGGCU--UUAGUGCUUUUAUUAGCAAGUGCGCGAAAUUCGAUUUCGAU ..............(((---(.((((((((((....--...-------.)))))).))))..)))).--...(..(((........)))..).((((((...)))))).. ( -27.10) >DroYak_CAF1 105303 95 + 1 AAAAACCAAAUCCCGCC----CCAAUGGCCAUAAAA--CUC-------GGUGGCCAUUUGUUGGGCU--UUAGUACUUUUAUUAGCAAGUGCGCGAAAUUCGAUUUCCAU .......(((((..(((----(.(((((((((....--...-------.)))))))))....)))).--...((((((........)))))).........))))).... ( -26.90) >consensus AAAAACCAAAUCCCGCC___CCCAAAGGCCAUAAAA__CGC_______GGUGGCCAUUUGUCGGGCU__UUAGUAUUUUUAUUAGCAAGUACGCGAAAUUCGAUUUCCAU ..............(((.....((((((((((.................)))))).))))...)))......((((((........)))))).................. (-14.70 = -13.93 + -0.77)

| Location | 14,639,197 – 14,639,295 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.62 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -15.77 |
| Energy contribution | -17.53 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
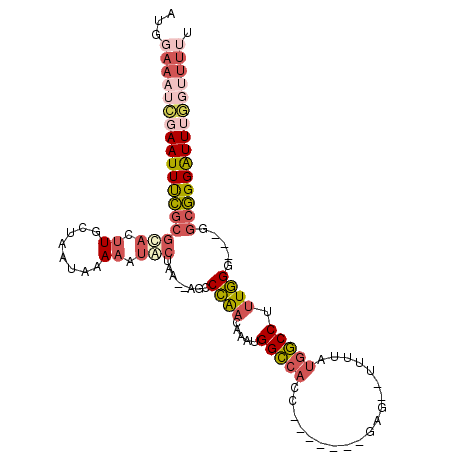
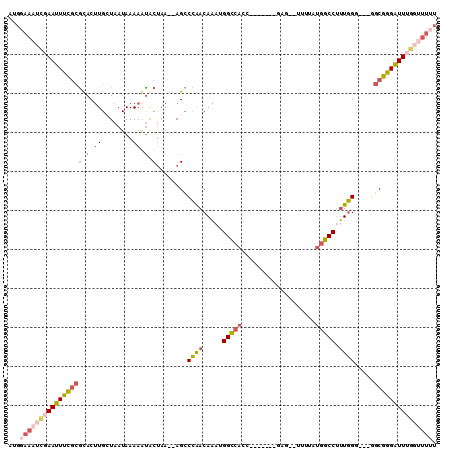
>X_DroMel_CAF1 14639197 98 - 22224390 AUGGAAAUCGAAUUUCGCGUACUUGCUAAUAAAAAUACUAA--AGCCCGACAAAUGGCCACC-------GCCGUUUUUAUGGCCUUUGGG---AGCGGGAUUUGGUUUUU ..(((((((((((..(((((....))...........((((--((.(((..(((((((....-------)))))))...))).)))))).---.)))..))))))))))) ( -32.40) >DroGri_CAF1 117577 103 - 1 AUGAAACUGAAAUUCA--ACACAUGCUAAUAAAAAUGCCCAUGAGACAAACAUCUGGACAAUCCAUGUCUAG--CUUCAGUUCCUAUUGG---AUCUGGGUUGCCUUUGU .........(((..((--((.((((...((....))...))))..........(((((((.....)))))))--..((((.(((....))---).))))))))..))).. ( -18.40) >DroSec_CAF1 109003 92 - 1 AUGGAAAUCGAAUUUCGCGUACUUGAUAAUAAAAAUACUAA--AGCCCGACAAAUGGCCACC-------GCG--UUUUAUGGCCUUUGGUGGUGGCGGGAUU-------U .........((((..((((((.((........)).)))...--.(((...((((.(((((..-------...--.....)))))))))..))).)))..)))-------) ( -22.50) >DroSim_CAF1 106756 99 - 1 AUGGAAAUCGAAUUUCGCGUACUUGAUAAUAAAAAUACUAA--AGCCCGACAAAUGGCCACC-------GCG--UUUUAUGGCCUUUGGUGGUGGCGGGAUUUGGUUUUU ..(((((((((((..((((((.((........)).)))...--.(((...((((.(((((..-------...--.....)))))))))..))).)))..))))))))))) ( -30.30) >DroEre_CAF1 102712 96 - 1 AUCGAAAUCGAAUUUCGCGCACUUGCUAAUAAAAGCACUAA--AGCCCAACAAAUGGCCACC-------AAG--UUUUAUGGCCUUUGGG---GGCGGGAUUUGGUUUUU ...((((((((((..(((.....((((......))))....--..(((..((((.(((((..-------...--.....)))))))))))---))))..)))))))))). ( -33.10) >DroYak_CAF1 105303 95 - 1 AUGGAAAUCGAAUUUCGCGCACUUGCUAAUAAAAGUACUAA--AGCCCAACAAAUGGCCACC-------GAG--UUUUAUGGCCAUUGG----GGCGGGAUUUGGUUUUU ..(((((((((((..(((..((((........)))).....--..((((....(((((((..-------...--.....))))))))))----))))..))))))))))) ( -33.30) >consensus AUGGAAAUCGAAUUUCGCGCACUUGCUAAUAAAAAUACUAA__AGCCCAACAAAUGGCCACC_______GAG__UUUUAUGGCCUUUGGG___GGCGGGAUUUGGUUUUU ...((((((((((((((((((.((........)).)))........((((.....(((((...................))))).)))).....))))))))))))))). (-15.77 = -17.53 + 1.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:27 2006