| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,605,296 – 14,605,528 |
| Length | 232 |
| Max. P | 0.877547 |

| Location | 14,605,296 – 14,605,411 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.10 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -18.92 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14605296 115 + 22224390 AACUUGGGGCCAUGGAAGACAAUAAAUAUUUAUAUGUAAAUGCCGGCAAUAUUGUCU-CUAUU-GUGCACAC-AGGGCACCAAACAAACAAAAAAAAAAGAAGAAGGUGGG-GAU-CCUG ...((((.(((.((((.(((((((.((((....))))...((....)).))))))))-)))((-((....))-))))).)))).....................(((....-...-))). ( -21.60) >DroSec_CAF1 79761 118 + 1 AACUUGGGGCCAUGGAAGACAAUAAAUAUUUAUAUGUAAAUGCCAGCAAUAUUGUCUUCUAUUGGUGCACACCAGGGCACCAAACAAACAAAUAAGGA--AAAAAGGUGGGGGAUCCCAG ....(((((((.((((((((((((.((((....))))...((....)).))))))))))))(((((((........)))))))...............--...........)).))))). ( -33.80) >DroSim_CAF1 79826 112 + 1 AACUUGGGGCCAUGGAAGACAAUAAAUAUUUAUAUGUAAAUGCCAGCAAUAUUGUCU-CUAUU-GUGCACAC-AGGGCACCAAACAAACAAACAAA-A--AAAAAGGUGGG-GAU-CCAG ...((((.(((.((((.(((((((.((((....))))...((....)).))))))))-)))((-((....))-))))).)))).............-.--.......(((.-...-))). ( -21.60) >DroEre_CAF1 74631 93 + 1 AACUUGGGGCCAUGGAAGACAAUAAAUAUUUAUAUGUAAAUGCCGGCAAUAUUGUCU-CUAUU-GUG-GCAC-AGGGCACCA---------------------AAGGUGGG-CAU-UCUG ..((((..((((..(.((((((((.((((....))))...((....)).))))))))-...).-.))-)).)-)))((.(((---------------------....))))-)..-.... ( -24.90) >DroYak_CAF1 75979 111 + 1 AACUUGGGGCCAUGGAAGACAAUAAAUAUUUAUAUGUAAAUGCCGGCAAUAUUGUCU-UCAUU-GUGCACAC-AGGGCACCAAACAAACAAACGCA----AAAAAGGUGGA-GAU-UUUG ...((((.(((...((((((((((.((((....))))...((....)).))))))))-)).((-((....))-))))).)))).((((....(((.----......)))..-...-)))) ( -24.30) >consensus AACUUGGGGCCAUGGAAGACAAUAAAUAUUUAUAUGUAAAUGCCGGCAAUAUUGUCU_CUAUU_GUGCACAC_AGGGCACCAAACAAACAAACAAA_A__AAAAAGGUGGG_GAU_CCUG ...((((.(((.(((.((((((((.((((....))))...((....)).)))))))).)))(((((....)))))))).))))..................................... (-18.92 = -19.00 + 0.08)


| Location | 14,605,373 – 14,605,489 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.10 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -21.92 |
| Energy contribution | -22.76 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14605373 116 + 22224390 CAAACAAACAAAAAAAAAAGAAGAAGGUGGG-GAU-CCUGCUAACCUGAGUC-CGGAGGGUACAGUUCACACCAUGCUAAGUGCGCCGG-AGGUCGCACUAAAGUAAGCCAUUAAAUGUA .......(((...............((((.(-(((-..((((..((......-.))..))))..)))).)))).((((.((((((((..-.)).))))))..))))..........))). ( -27.10) >DroSec_CAF1 79841 118 + 1 CAAACAAACAAAUAAGGA--AAAAAGGUGGGGGAUCCCAGCUAACCUGAGCCCCGGAGGGUACAGUUCACACCCUGCUAAGUGCGCCGGAAGGUCGCACUAAAGUAAGCCAUUAAAUGUA .......(((..(((((.--.....(((((((...))).......(((.((((....)))).)))....)))).((((.((((((((....)).))))))..))))..)).)))..))). ( -36.20) >DroSim_CAF1 79903 113 + 1 CAAACAAACAAACAAA-A--AAAAAGGUGGG-GAU-CCAGCUAACCUGAGCC-CGGAGGGUACAGUUUACACCAUGCUAAGUGCGCCGG-AGGUCGCACUAAAGUAAGCCAUUAAAUGUA ...........(((..-.--.....((((((-...-))...(((.(((.(((-(...)))).))).))))))).((((.((((((((..-.)).))))))..))))..........))). ( -26.20) >DroEre_CAF1 74707 95 + 1 CA---------------------AAGGUGGG-CAU-UCUGCUGACCUGAACC-CGCAGGGCACAGUUCACACCAUGCUAAGUGCACCGG-AGGUCGCACUAAAGUAAGCCAUUAAGUGUA ..---------------------..(((.((-(((-..((.(((.(((..((-(...)))..))).)))))..))))).((((((((..-.))).))))).......))).......... ( -26.60) >DroYak_CAF1 76056 112 + 1 CAAACAAACAAACGCA----AAAAAGGUGGA-GAU-UUUGCUAACCUAAACC-CGGAGGGUACAGUUCACACCAUGCUAAGUGCGCCAG-AGGUCGCACUAAAGUAAGACAUUAAAUGUA .............(((----.....((((..-(((-(.((((..((......-.))..)))).))))..)))).((((.((((((((..-.)).))))))..))))..........))). ( -27.50) >consensus CAAACAAACAAACAAA_A__AAAAAGGUGGG_GAU_CCUGCUAACCUGAGCC_CGGAGGGUACAGUUCACACCAUGCUAAGUGCGCCGG_AGGUCGCACUAAAGUAAGCCAUUAAAUGUA .........................((((...(((...((((..((........))..))))..)))..)))).((((.((((((((....)).))))))..)))).............. (-21.92 = -22.76 + 0.84)



| Location | 14,605,411 – 14,605,528 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.26 |
| Mean single sequence MFE | -37.14 |
| Consensus MFE | -26.24 |
| Energy contribution | -27.84 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14605411 117 + 22224390 CUAACCUGAGUC-CGGAGGGUACAGUUCACACCAUGCUAAGUGCGCCGG-AGGUCGCACUAAAGUAAGCCAUUAAAUGUAACAAU-UGAUAUAAUUGUACGCAUUAUUUGGGCUGCAGGA ....((((((((-((((((.((((((.........))).((((((((..-.)).))))))...)))..))....(((((.(((((-((...)))))))..))))).)))))))).)))). ( -35.50) >DroSec_CAF1 79879 120 + 1 CUAACCUGAGCCCCGGAGGGUACAGUUCACACCCUGCUAAGUGCGCCGGAAGGUCGCACUAAAGUAAGCCAUUAAAUGUAACAAUUUGAUAUAAUUGUACCCAUUGCUUUGGCUGCAGGA ....((((((((..((.((((((((((.......((((.((((((((....)).))))))..))))....(((((((......)))))))..))))))))))....))..)))).)))). ( -46.40) >DroSim_CAF1 79938 117 + 1 CUAACCUGAGCC-CGGAGGGUACAGUUUACACCAUGCUAAGUGCGCCGG-AGGUCGCACUAAAGUAAGCCAUUAAAUGUAACAAU-UGAUAUAAUUGUACCCAUUGCUUGGGCUGCAGGA ....((((((((-(((.((((((((((.......((((.((((((((..-.)).))))))..))))....(((((.((...)).)-))))..))))))))))....).)))))).)))). ( -41.00) >DroEre_CAF1 74724 116 + 1 CUGACCUGAACC-CGCAGGGCACAGUUCACACCAUGCUAAGUGCACCGG-AGGUCGCACUAAAGUAAGCCAUUAAGUGUAACAAU-UGACAUAAUUGUAUCCAUUGCGUGUGCUGCGGA- ...........(-(((((.((((...........((((.((((((((..-.))).)))))..)))).((.....((((..(((((-((...)))))))...)))))))))).)))))).- ( -34.00) >DroYak_CAF1 76090 116 + 1 CUAACCUAAACC-CGGAGGGUACAGUUCACACCAUGCUAAGUGCGCCAG-AGGUCGCACUAAAGUAAGACAUUAAAUGUAACAAU-UGAUAUAAUUGUACCCAUUGCGUGUGCUGCAGC- ....((......-.)).((((((((((.......((((.((((((((..-.)).))))))..))))..((((...))))......-......)))))))))).(((((.....))))).- ( -28.80) >consensus CUAACCUGAGCC_CGGAGGGUACAGUUCACACCAUGCUAAGUGCGCCGG_AGGUCGCACUAAAGUAAGCCAUUAAAUGUAACAAU_UGAUAUAAUUGUACCCAUUGCUUGGGCUGCAGGA ....((((((((.(((.((((((((((.......((((.((((((((....)).))))))..)))).........((((.........)))))))))))))).)))....)))).)))). (-26.24 = -27.84 + 1.60)


| Location | 14,605,411 – 14,605,528 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.26 |
| Mean single sequence MFE | -35.72 |
| Consensus MFE | -28.60 |
| Energy contribution | -29.04 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
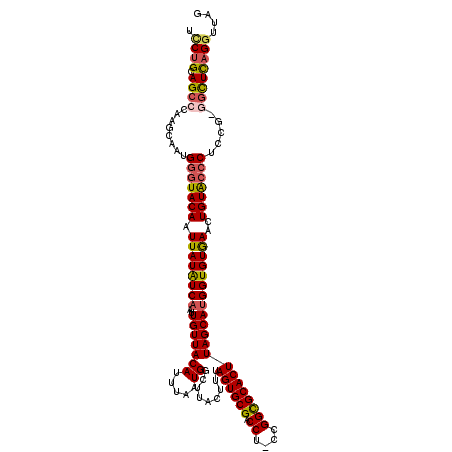
>X_DroMel_CAF1 14605411 117 - 22224390 UCCUGCAGCCCAAAUAAUGCGUACAAUUAUAUCA-AUUGUUACAUUUAAUGGCUUACUUUAGUGCGACCU-CCGGCGCACUUAGCAUGGUGUGAACUGUACCCUCCG-GACUCAGGUUAG .((((.((.((.....((((((((((((.....)-)))).))).................((((((.((.-..))))))))..))))((.(((.....)))))...)-).)))))).... ( -28.30) >DroSec_CAF1 79879 120 - 1 UCCUGCAGCCAAAGCAAUGGGUACAAUUAUAUCAAAUUGUUACAUUUAAUGGCUUACUUUAGUGCGACCUUCCGGCGCACUUAGCAGGGUGUGAACUGUACCCUCCGGGGCUCAGGUUAG (((((((((((....((((...((((((......))))))..))))...)))))......((((((.((....))))))))..))))))......(((..(((....)))..)))..... ( -41.00) >DroSim_CAF1 79938 117 - 1 UCCUGCAGCCCAAGCAAUGGGUACAAUUAUAUCA-AUUGUUACAUUUAAUGGCUUACUUUAGUGCGACCU-CCGGCGCACUUAGCAUGGUGUAAACUGUACCCUCCG-GGCUCAGGUUAG .((((.(((((.......(((((((.((((((((-.......(((...)))(((......((((((.((.-..)))))))).))).))))))))..)))))))...)-)))))))).... ( -42.10) >DroEre_CAF1 74724 116 - 1 -UCCGCAGCACACGCAAUGGAUACAAUUAUGUCA-AUUGUUACACUUAAUGGCUUACUUUAGUGCGACCU-CCGGUGCACUUAGCAUGGUGUGAACUGUGCCCUGCG-GGUUCAGGUCAG -(((((((..((((((((.((((......)))).-)))))((((((.....(((......(((((.(((.-..)))))))).)))..))))))....)))..)))))-)).......... ( -36.40) >DroYak_CAF1 76090 116 - 1 -GCUGCAGCACACGCAAUGGGUACAAUUAUAUCA-AUUGUUACAUUUAAUGUCUUACUUUAGUGCGACCU-CUGGCGCACUUAGCAUGGUGUGAACUGUACCCUCCG-GGUUUAGGUUAG -.(((.(((...((....(((((((.((((((((-..((((((((...))).........((((((.((.-..)))))))))))))))))))))..)))))))..))-.))))))..... ( -30.80) >consensus UCCUGCAGCCCAAGCAAUGGGUACAAUUAUAUCA_AUUGUUACAUUUAAUGGCUUACUUUAGUGCGACCU_CCGGCGCACUUAGCAUGGUGUGAACUGUACCCUCCG_GGCUCAGGUUAG .((((.((((........(((((((.((((((((...(((((((.....)).........((((((.((....)))))))))))))))))))))..))))))).....)))))))).... (-28.60 = -29.04 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:18 2006