
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,604,815 – 14,604,962 |
| Length | 147 |
| Max. P | 0.966145 |

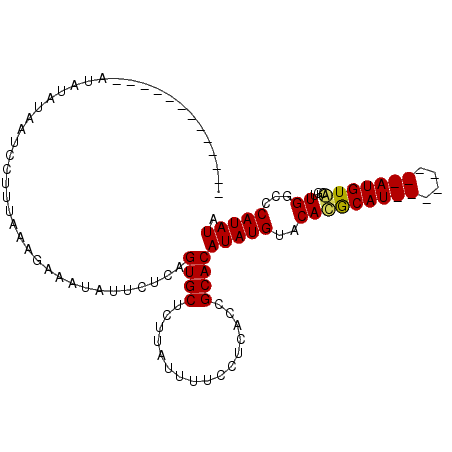
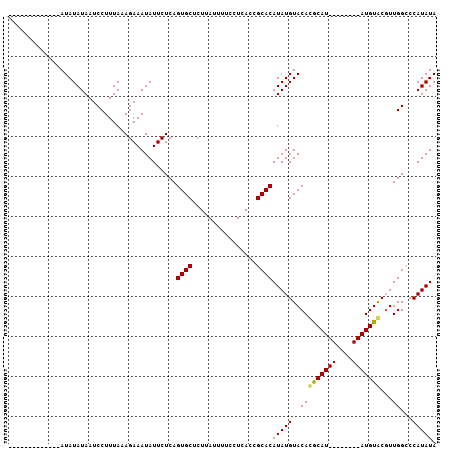
| Location | 14,604,815 – 14,604,922 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.00 |
| Mean single sequence MFE | -16.09 |
| Consensus MFE | -9.60 |
| Energy contribution | -9.82 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.880992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14604815 107 + 22224390 AUAUAUAACCUUUAUAUAUAAUCCUUUAAAGAAAUAUUCUCAGUGCUCUUAUUUUCCUCACUGCACAUAUGUACAUACAUACAUAUGUAUGUACGUUGGCCCAUAUA (((((((.....)))))))...((..................((((................))))..((((((((((((....)))))))))))).))........ ( -20.19) >DroSec_CAF1 79310 86 + 1 -------------AUAUAUAAUCCUUUAAAGAAAUAUUCUCAGUGCUCUUAUUUUCCUCACCGCACAUAUGUACACGCAU--------AUGUACUUUGGCCCAUAUA -------------............((((((...........((((................))))((((((....))))--------))...))))))........ ( -9.89) >DroSim_CAF1 79368 86 + 1 -------------AUAUAUAAUCCUUUAAAGAAAUAUUCUCAGUGCUCUUAUUUUCCUCACCGCACAUAUGUACACGCAU--------AUGUGCGUUGGUCCAUAUA -------------.(((((...((....((((.((.......))..))))...........(((((((((((....))))--------)))))))..))...))))) ( -18.20) >consensus _____________AUAUAUAAUCCUUUAAAGAAAUAUUCUCAGUGCUCUUAUUUUCCUCACCGCACAUAUGUACACGCAU________AUGUACGUUGGCCCAUAUA ..........................................((((................))))(((((..(((((((((....)))))))...))...))))). ( -9.60 = -9.82 + 0.23)

| Location | 14,604,815 – 14,604,922 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.00 |
| Mean single sequence MFE | -23.09 |
| Consensus MFE | -14.83 |
| Energy contribution | -14.72 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14604815 107 - 22224390 UAUAUGGGCCAACGUACAUACAUAUGUAUGUAUGUACAUAUGUGCAGUGAGGAAAAUAAGAGCACUGAGAAUAUUUCUUUAAAGGAUUAUAUAUAAAGGUUAUAUAU (((((((.((...((((((((((....))))))))))((((((((((((.............))))).......((((....)))).)))))))...))))))))). ( -25.42) >DroSec_CAF1 79310 86 - 1 UAUAUGGGCCAAAGUACAU--------AUGCGUGUACAUAUGUGCGGUGAGGAAAAUAAGAGCACUGAGAAUAUUUCUUUAAAGGAUUAUAUAU------------- (((((((.((...((((((--------(((......)))))))))..((((((((..................))))))))..)).))))))).------------- ( -19.77) >DroSim_CAF1 79368 86 - 1 UAUAUGGACCAACGCACAU--------AUGCGUGUACAUAUGUGCGGUGAGGAAAAUAAGAGCACUGAGAAUAUUUCUUUAAAGGAUUAUAUAU------------- (((((((.((..(((((((--------(((......)))))))))).((((((((..................))))))))..)).))))))).------------- ( -24.07) >consensus UAUAUGGGCCAACGUACAU________AUGCGUGUACAUAUGUGCGGUGAGGAAAAUAAGAGCACUGAGAAUAUUUCUUUAAAGGAUUAUAUAU_____________ (((((((.((...((((((............))))))((((...(((((.............)))))...)))).........)).))))))).............. (-14.83 = -14.72 + -0.11)

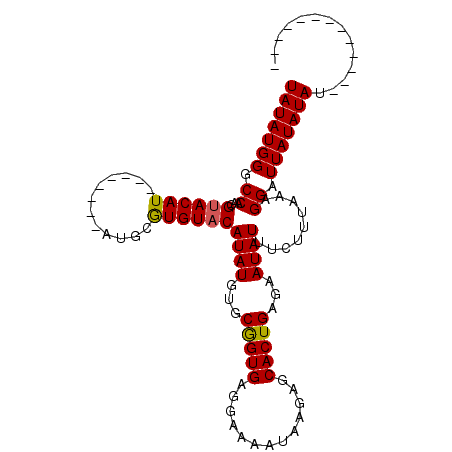
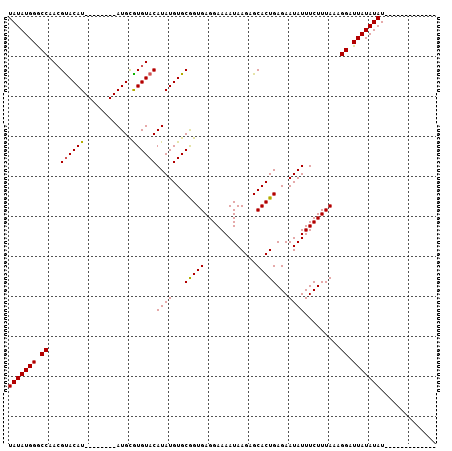
| Location | 14,604,855 – 14,604,962 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -26.04 |
| Consensus MFE | -10.58 |
| Energy contribution | -10.06 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.41 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14604855 107 - 22224390 GUCCUCCGGCUUUAUUAUUUCAUCAUUAUAACAUUUCCUC----UAUAUGGGCCAACGUACAUACAUAUGUAUGUAUGUAC------AUAUGUGCAGUG---AGGAAAAUAAGAGCACUG ........(((((.((((.........))))..(((((((----....((.((....((((((((((....))))))))))------....)).))..)---))))))...))))).... ( -27.50) >DroSec_CAF1 79337 99 - 1 GUCCUCCGGCUUUAUUAUUUCAUCAUUAUAACAUUUCCUC----UAUAUGGGCCAAAGUACAU--------AUGCGUGUAC------AUAUGUGCGGUG---AGGAAAAUAAGAGCACUG .(((((((((((...(((......................----.))).)))))...((((((--------(((......)------)))))))).).)---)))).............. ( -22.70) >DroSim_CAF1 79395 99 - 1 GUCCUCCGGCUUUAUUAUUUCAUCAUUAUAACAUUUCCUC----UAUAUGGACCAACGCACAU--------AUGCGUGUAC------AUAUGUGCGGUG---AGGAAAAUAAGAGCACUG ........(((((....((((.((((.........(((..----.....)))....(((((((--------(((......)------))))))))))))---).))))...))))).... ( -26.70) >DroEre_CAF1 74256 86 - 1 GUCCUCUGGCUUUAUUAUUUCAUCAUUAUAACAUUUCCC-----UAUUUGGGGCA-------------------CAUGUAC------AUAUGUGCGGUG----GGGAAAUAAGAGCGUUG ........((((((((.(..((((............(((-----.....)))(((-------------------((((...------.)))))))))))----..).))).))))).... ( -25.50) >DroYak_CAF1 75555 112 - 1 GUCCUCGGGCUUUAUUAUUUCAUCAUUAUAACAUUUCCUUUAUAUAUAUGGGGCAACUUACAU--------AUGCGUGUAUAUAUUGAUAUGUGCCGUGCGAAGGAAAAUAAGAGCACCG .....((((((((.((((.........))))..((((((((..((((.((((....)))).))--------))(((.(((((((....))))))))))..))))))))...))))).))) ( -27.80) >consensus GUCCUCCGGCUUUAUUAUUUCAUCAUUAUAACAUUUCCUC____UAUAUGGGCCAACGUACAU________AUGCGUGUAC______AUAUGUGCGGUG___AGGAAAAUAAGAGCACUG ........((((((((.((((..............(((...........))).....................(((..((........))..)))........))))))).))))).... (-10.58 = -10.06 + -0.52)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:15 2006