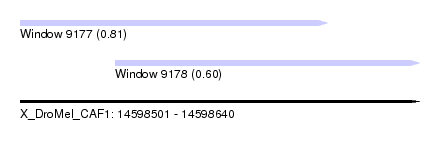
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,598,501 – 14,598,640 |
| Length | 139 |
| Max. P | 0.808417 |

| Location | 14,598,501 – 14,598,608 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 71.18 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -7.54 |
| Energy contribution | -8.21 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
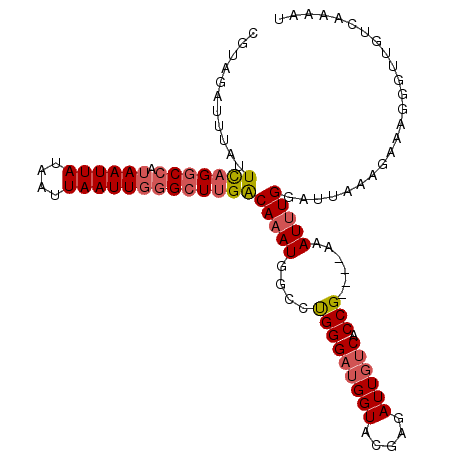
>X_DroMel_CAF1 14598501 107 + 22224390 CAACCAAAUUUAGUAUAAAUUAAAUGCCAU--UUGCGUAGAUUUAUUCAGGCCAUAAUUAUAAUUAAUUGGGCUUGACAAAUGGCCUGGGAU-GGUACGAGAUUGUCACC--- ...(((.(((((((....)))))))(((((--(((...........(((((((.((((((....))))))))))))))))))))).)))((.-.((.....))..))...--- ( -29.60) >DroPse_CAF1 85603 98 + 1 ---------UCAGAAGAGG----AGACUGU--UUUACUAAAUUGACUUAACUAAUAAUUAUAAUUAAUUCGGCUUGACAAAUGGCUUGGAAUUUGUGCGAGUAUCCCACGAGU ---------..((((.((.----...)).)--)))...............................((((((((((((((((........)))))).)))))......))))) ( -16.20) >DroSim_CAF1 73750 107 + 1 CAACCAAAUUUAGUAUAAAUUAAAUGCCAU--UUGCGUAGAUUUAUUCAGGCCAUAAUUAUAAUUAAUUGGGCUUGACAAAUGGCCUGGGAU-GGUACGAGAUUGUCACC--- ...(((.(((((((....)))))))(((((--(((...........(((((((.((((((....))))))))))))))))))))).)))((.-.((.....))..))...--- ( -29.60) >DroEre_CAF1 69658 96 + 1 CAACCAAAUUUAGAAUAAAUGAAUUGCCAU--UUGCGUAGAUUUAUUCAGGCCAUAAUUAUAAUUAAUUGGGCUUGACAAAUGGCCUGGGAU------------GUCACC--- ...(((((((((.......))))))(((((--(((...........(((((((.((((((....))))))))))))))))))))).)))...------------......--- ( -25.90) >DroYak_CAF1 70200 107 + 1 CAACCAAAUUUAGAAUAAAUGAAAUACCAU--UUGAGUCGAUUUAUUCAGGCCAUAAUUAUAAUUAAUUGGGCUUGACAAAUGGCUUGGGAU-GGUACGAGAUUGCCACC--- ........................((((((--(..((((.((((..(((((((.((((((....))))))))))))).))))))))..).))-)))).............--- ( -27.20) >DroAna_CAF1 109437 97 + 1 CAACCGAGUGUGGAAUAAAUUAAAUGCGAGAGUGGUUCAAAUUGUGUUCGCCAAUAAUUAUAAUUAAUUUAGCUUGGCAAAUCACUCGGGA-------------AUCACC--- ...(((((((..(((((((((...(((....))).....)))).)))))(((((.(((((....)))))....)))))....)))))))..-------------......--- ( -24.40) >consensus CAACCAAAUUUAGAAUAAAUUAAAUGCCAU__UUGCGUAGAUUUAUUCAGGCCAUAAUUAUAAUUAAUUGGGCUUGACAAAUGGCCUGGGAU_GGUACGAGAUUGUCACC___ .........................(((((................(((((((.((((((....)))))))))))))...)))))............................ ( -7.54 = -8.21 + 0.67)


| Location | 14,598,534 – 14,598,640 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.45 |
| Mean single sequence MFE | -23.56 |
| Consensus MFE | -14.38 |
| Energy contribution | -15.60 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14598534 106 + 22224390 CGUAGAUUUAUUCAGGCCAUAAUUAUAAUUAAUUGGGCUUGACAAAUGGCCUGGGAUGGUACGAGAUUGUCACCG----AAAUUUGGAUUAAAGAAAGGGUUGUCGAAAU ...........(((((((.((((((....)))))))))))))...(..((((..((..((.....))..)).(((----(...))))..........))))..)...... ( -23.70) >DroSim_CAF1 73783 106 + 1 CGUAGAUUUAUUCAGGCCAUAAUUAUAAUUAAUUGGGCUUGACAAAUGGCCUGGGAUGGUACGAGAUUGUCACCG----AAAUUUGGAUUAAAGAAAGGGUUGUCGAAAU ...........(((((((.((((((....)))))))))))))...(..((((..((..((.....))..)).(((----(...))))..........))))..)...... ( -23.70) >DroEre_CAF1 69691 93 + 1 CGUAGAUUUAUUCAGGCCAUAAUUAUAAUUAAUUGGGCUUGACAAAUGGCCUGGGAU-----------GUCACCG----AAAUUUGUAUUUAAGUAAGGGUGG--AAAAU ..........(((((((((((((((....))))).(......)..))))))))))..-----------.(((((.----..(((((....)))))...)))))--..... ( -21.00) >DroYak_CAF1 70233 106 + 1 AGUCGAUUUAUUCAGGCCAUAAUUAUAAUUAAUUGGGCUUGACAAAUGGCUUGGGAUGGUACGAGAUUGCCACCG----AAAUCUGGAUUAAAGAAAGGGUUGUCAAAAU .(.((((((..(((((((.((((((....))))))))))))).....(..((.((.(((((......))))))).----))..).............)))))).)..... ( -26.00) >DroAna_CAF1 109472 92 + 1 UUCAAAUUGUGUUCGCCAAUAAUUAUAAUUAAUUUAGCUUGGCAAAUCACUCGGGA------------AUCACCGCCAAAAAUUUGGAUGGAAUU----GGAC--ACAAA ......(((((((((((((.(((((....)))))....))))).............------------....(((((((....)))).)))....----))))--)))). ( -23.40) >consensus CGUAGAUUUAUUCAGGCCAUAAUUAUAAUUAAUUGGGCUUGACAAAUGGCCUGGGAUGGUACGAGAUUGUCACCG____AAAUUUGGAUUAAAGAAAGGGUUGUCAAAAU ...........(((((((.((((((....)))))))))))))(((((....(((((((((.....)))))).)))......)))))........................ (-14.38 = -15.60 + 1.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:12 2006