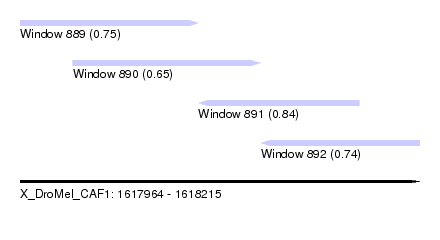
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,617,964 – 1,618,215 |
| Length | 251 |
| Max. P | 0.840707 |

| Location | 1,617,964 – 1,618,076 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.13 |
| Mean single sequence MFE | -36.11 |
| Consensus MFE | -24.15 |
| Energy contribution | -23.79 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1617964 112 + 22224390 AGCAGCACCCAUCAUUAAAUCGCUGCAUUUCG-------CUUGCGAAAUGGCCAUUUAAUCGGCCAUCCUGUGGCUUCUGGGUCACCAUUUUGCUAGCUGUGUAUGG-CAAGCCGAAUAU .(((((...............))))).....(-------((.(((((((((((........)))).....((((((....)))))).))))))).))).((((.(((-....))).)))) ( -35.36) >DroSec_CAF1 13636 112 + 1 AGCAGCUCCCAUCAUUAAAUCGCAGCACUUCG-------CUAGCGAAAUGGCCAUUUAAUCGGCCAUCCUGUGGCUUCUGGGUCACCAUUUUACUAGCUGUGUAUGGGCAAGCC-AAUAA ....((((((((........((((((..((((-------....))))((((((........))))))...((((((....))))))..........)))))).)))))..))).-..... ( -34.10) >DroSim_CAF1 29556 112 + 1 AGCAGCUCCCAUCAUUAAAUCGCAGCACUUCG-------CUAGCGAAAUGGCCAUUUAAUCGGCCAUCCUGUGGCUUCUGGGUCACCAUUUUGCUAGCUGUGUAUGGGCAAGCC-AAUAG ....((((((((............((((...(-------((((((((((((((........)))).....((((((....)))))).))))))))))).)))))))))..))).-..... ( -39.30) >DroEre_CAF1 14134 112 + 1 AGCAGCUCACAUCAUUAAAUCGCAGCACUGCG-------CCAGUGAAAUGGCCAUUUAAUCGGCCGUCCUGUGGGUUCUGGGUCACCAAUAUGCCAGCUGCGGCCAGCCAAGG-GAACGU .(((((((((((((((....((((....))))-------..))))).((((((........))))))..))))(((..(((....)))....)))))))))((....))..(.-...).. ( -37.50) >DroYak_CAF1 15495 118 + 1 AGCAGCUCACAUCAUUAAAUCGCAGCAUUUCGUAGCAGUCAAAUGGAAUGGCCAUUUAAUCGGCCAUCCUGUGGCUUCUGGGUCACCA-UUUGCCAGCUGUGGGUCACCAUCG-CCACAU ....................((((((......(((.(((((...(((.(((((........))))))))..))))).)))(((.....-...))).))))))(((.......)-)).... ( -34.30) >consensus AGCAGCUCCCAUCAUUAAAUCGCAGCACUUCG_______CUAGCGAAAUGGCCAUUUAAUCGGCCAUCCUGUGGCUUCUGGGUCACCAUUUUGCUAGCUGUGUAUGGCCAAGCC_AAUAU .(((((.............((((...................)))).((((((........))))))....((((...(((....)))....)))))))))................... (-24.15 = -23.79 + -0.36)

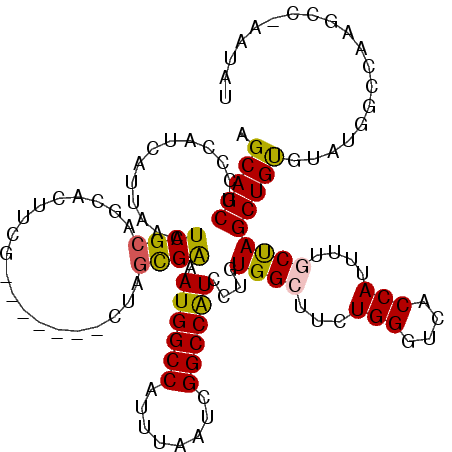
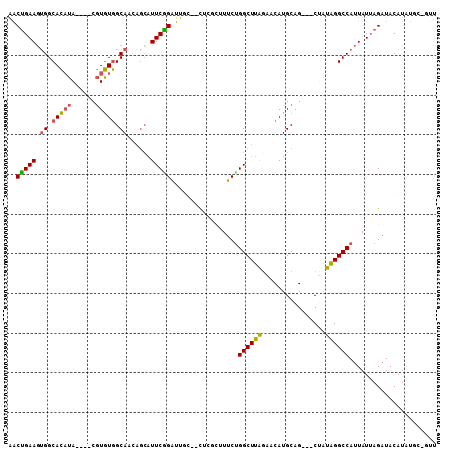
| Location | 1,617,997 – 1,618,115 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.60 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -19.70 |
| Energy contribution | -18.94 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1617997 118 + 22224390 UUGCGAAAUGGCCAUUUAAUCGGCCAUCCUGUGGCUUCUGGGUCACCAUUUUGCUAGCUGUGUAUGG-CAAGCCGAAUAUAAC-GAAUAUGUAUCUAAUAAUGGCCUAUAGAUGCUGCAU .......((((((........))))))..((..((.(((((((((.......(((.(((......))-).))).(((((((..-...))))).))......))))))..))).))..)). ( -32.40) >DroSec_CAF1 13669 115 + 1 UAGCGAAAUGGCCAUUUAAUCGGCCAUCCUGUGGCUUCUGGGUCACCAUUUUACUAGCUGUGUAUGGGCAAGCC-AAUAAAAC-GAAUAUGUAUCUAAUAAUGGCCUAUAG---CUGCAU .......((((((........))))))..((..(((..(((((((..(((.(((((..(((.....((....))-......))-)..)).)))...)))..))))))).))---)..)). ( -31.10) >DroSim_CAF1 29589 115 + 1 UAGCGAAAUGGCCAUUUAAUCGGCCAUCCUGUGGCUUCUGGGUCACCAUUUUGCUAGCUGUGUAUGGGCAAGCC-AAUAGAAC-GUAUAUGUAUCUAAUAAUGGCCUAUAG---CUGCAU (((((((((((((........)))).....((((((....)))))).)))))))))((.((.(((((((.....-..((((((-(....))).))))......))))))))---).)).. ( -37.12) >DroEre_CAF1 14167 116 + 1 CAGUGAAAUGGCCAUUUAAUCGGCCGUCCUGUGGGUUCUGGGUCACCAAUAUGCCAGCUGCGGCCAGCCAAGG-GAACGUAGAUACAAAUGUAUCUAAUAAUGGCCCAAUG---CUGCAU ((((.....(((((((.....((((((.(((.(.((..(((....)))..)).))))..))))))........-.....(((((((....)))))))..)))))))....)---)))... ( -40.30) >DroYak_CAF1 15535 114 + 1 AAAUGGAAUGGCCAUUUAAUCGGCCAUCCUGUGGCUUCUGGGUCACCA-UUUGCCAGCUGUGGGUCACCAUCG-CCACAUGGA-GCAGAUGUAUCUUAUAUUGGCCUAUUG---CUGCAU ....(((.(((((........))))))))((..((...(((((((.((-(((((...((((((((.......)-)).))))).-)))))))..........)))))))..)---)..)). ( -40.00) >consensus UAGCGAAAUGGCCAUUUAAUCGGCCAUCCUGUGGCUUCUGGGUCACCAUUUUGCUAGCUGUGUAUGGCCAAGCC_AAUAUAAC_GAAUAUGUAUCUAAUAAUGGCCUAUAG___CUGCAU .......((((((........))))))..((..(....(((((((.......((.....))((....))................................)))))))......)..)). (-19.70 = -18.94 + -0.76)

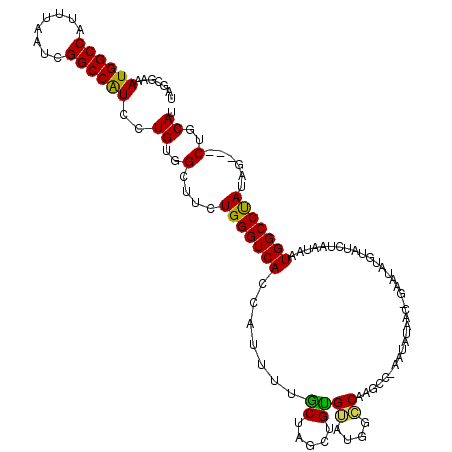
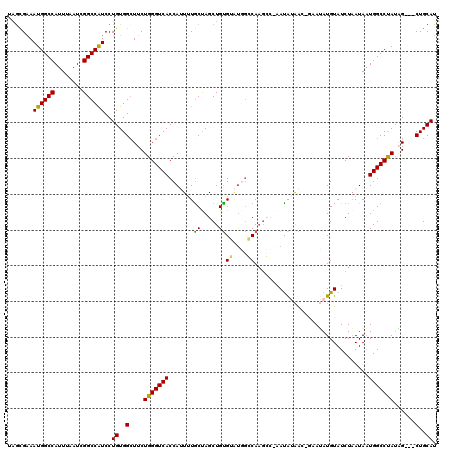
| Location | 1,618,076 – 1,618,177 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 74.66 |
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -14.47 |
| Energy contribution | -14.19 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1618076 101 - 22224390 AACUGAAGUGGCACAUA----CGUGUGGCAACAGCAUUCGGAUUGC--CUAGCUUUCCGGCUUAGAACAUGCAGCAUCUAUAGGCCAUUAUUAGAUACAUAUUC-GUU ..((((((((((..(((----.(((((....).(((((((((..((--...))..)))))........)))).)))).)))..)))))).))))..........-... ( -26.50) >DroSec_CAF1 13748 98 - 1 AACUGAAGUGGCACAUA----CGUGUGGCAAAAGCAUUCGGAUUGC--CUGGAUUUCUGGCUUAGAACAUGCAG---CUAUAGGCCAUUAUUAGAUACAUAUUC-GUU ..((((((((((..(((----((((((....((((((((((.....--)))))).....))))....))))).)---.)))..)))))).))))..........-... ( -21.30) >DroSim_CAF1 29668 97 - 1 AACUGAAGUGGCACAUA----CGUGUGGCAACAGCAUUCGGAUUGC--C-GGCUUUCUGGCUUAGAACAUGCAG---CUAUAGGCCAUUAUUAGAUACAUAUAC-GUU ..((((((((((..(((----.((.((....))((((.......((--(-((....))))).......)))).)---))))..)))))).))))..........-... ( -24.64) >DroEre_CAF1 14246 100 - 1 GGCUGAAGUGGCACAUAUGCACGUGUGGCAC-----UUCAGCUUCUGAUGCUCAUGCUGGCUUAGGACAUGCAG---CAUUGGGCCAUUAUUAGAUACAUUUGUAUCU ((((((((((.(((((......))))).)))-----)))))))((..(((((((((((......)).)))).))---)))..))........((((((....)))))) ( -43.70) >DroYak_CAF1 15613 104 - 1 AACAGAAAAGGCAUCUACGCACCUAUAGCAUCAGCAUUCUGAUUCUGAUUCUUAUUCUGGCUUGCAACAUGCAG---CAAUAGGCCAAUAUAAGAUACAUCUGC-UCC ..((((....((......)).....(((.(((((....))))).)))..((((((..((((((((........)---))...)))))..))))))....)))).-... ( -23.90) >consensus AACUGAAGUGGCACAUA____CGUGUGGCAACAGCAUUCGGAUUGC__CUCGCUUUCUGGCUUAGAACAUGCAG___CUAUAGGCCAUUAUUAGAUACAUAUGC_GUU ..(((((.((.(((((......))))).))......))))).................((((((................))))))...................... (-14.47 = -14.19 + -0.28)

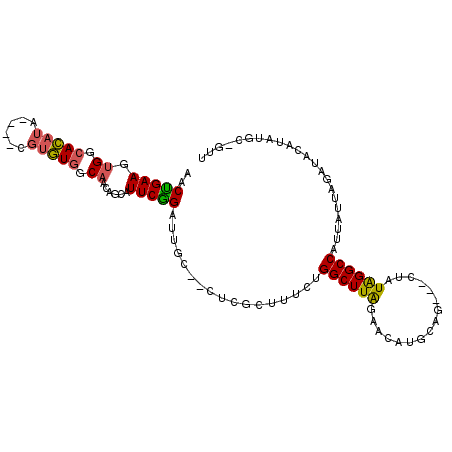
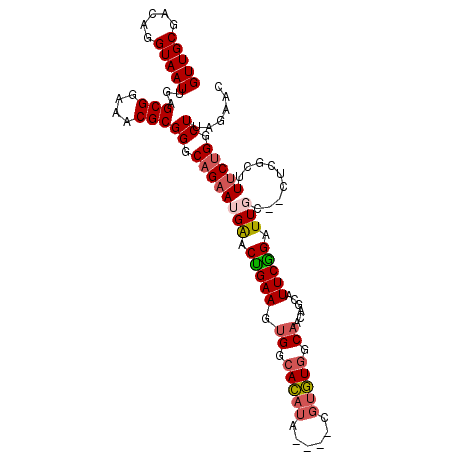
| Location | 1,618,115 – 1,618,215 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.00 |
| Mean single sequence MFE | -34.66 |
| Consensus MFE | -18.72 |
| Energy contribution | -19.40 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1618115 100 - 22224390 GUUGCGACAGGUAAUUGAGCGGAAACGCGGGCUGAAUGAACUGAAGUGGCACAUA----CGUGUGGCAACAGCAUUCGGAUUGC--CUAGCUUUCCGGCUUAGAAC (((((.....)))))((((((((((.((((((((((((..(((..((.((((...----.)))).))..)))))))))....))--)).))))))).))))).... ( -34.20) >DroSec_CAF1 13784 100 - 1 GUUGCGACAGGUAAUUGAGCGGAAACGCGGGCAGAAUGAACUGAAGUGGCACAUA----CGUGUGGCAAAAGCAUUCGGAUUGC--CUGGAUUUCUGGCUUAGAAC (((((.....)))))((((((((((..(((((((..((((.((...((.((((..----..)))).))....))))))..))))--)))..))))).))))).... ( -32.00) >DroSim_CAF1 29704 99 - 1 GUUGCGACAGGUAAUUGAGCGGAAACGCGGGCAGAAUGAACUGAAGUGGCACAUA----CGUGUGGCAACAGCAUUCGGAUUGC--C-GGCUUUCUGGCUUAGAAC (((((.....)))))((((((((((.((.(((((..(((((((..((.((((...----.)))).))..)))..))))..))))--)-.))))))).))))).... ( -35.30) >DroEre_CAF1 14283 101 - 1 GUUGCGACAGGUAAUUGAGCGGAAACGCGGACAGAAUGGGCUGAAGUGGCACAUAUGCACGUGUGGCAC-----UUCAGCUUCUGAUGCUCAUGCUGGCUUAGGAC ...((....((((..((((((....).....((((...((((((((((.(((((......))))).)))-----)))))))))))..))))))))).))....... ( -40.60) >DroYak_CAF1 15649 106 - 1 GUUGCGACAGGUAAUUGAGCGGAAACGCGGGCAGAAUGAACAGAAAAGGCAUCUACGCACCUAUAGCAUCAGCAUUCUGAUUCUGAUUCUUAUUCUGGCUUGCAAC (((((((...........(((....))).(.((((((((((((((...((......))..........((((....))))))))).)))..)))))).)))))))) ( -31.20) >consensus GUUGCGACAGGUAAUUGAGCGGAAACGCGGGCAGAAUGAACUGAAGUGGCACAUA____CGUGUGGCAACAGCAUUCGGAUUGC__CUCGCUUUCUGGCUUAGAAC (((((.....)))))...(((....)))((.((((((((.(((((.((.(((((......))))).))......))))).))).........))))).))...... (-18.72 = -19.40 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:55 2006