
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,592,121 – 14,592,232 |
| Length | 111 |
| Max. P | 0.976543 |

| Location | 14,592,121 – 14,592,232 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -46.13 |
| Consensus MFE | -23.62 |
| Energy contribution | -23.98 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
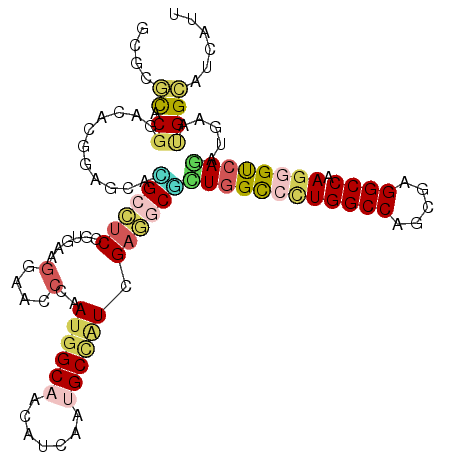
>X_DroMel_CAF1 14592121 111 + 22224390 GAUGAUCCCGUUCAUCUGACCCUUGGCCUCGCUGGCCAGGACCAACGCUUCGAUAGCAUUGAUAUUGCCAUUGGGAUCCUUCACGGCGGCACUUUCAGCAUCCGCGGAGGC ...(((((((...........(((((((.....)))))))..(((.(((.....))).)))..........)))))))((((.(((..((.......))..))).)))).. ( -37.40) >DroVir_CAF1 71837 111 + 1 AAUGAUGCCAUUCAGCUGGCCCUUGGCCUCGCUGGCCAGGGCCAGCGCCUCGACAGCAUUGAUGUUGCCAUUGGGCUCUCGUAUGGAGCCGUGCUCCGUGUCCGAGCGCGA ..(((......)))(((((((((.((((.....)))))))))))))(((((((.(((.(..(((....)))..)))).)).((((((((...))))))))...))).)).. ( -54.80) >DroPse_CAF1 78727 111 + 1 AAUGAUGCCAUUCAUUUGACCCUUGGCCUCACUGGCCAAGGCCAGAGCCUCGAUGGCGUUGAUGUUGCCAUUGGGAUCCUUGAUGGAGGCGCGUUCCGCAUCGGUACGCGC ......(((.((((((....((((((((.....))))))))...((..((((((((((.......)))))))))).))...)))))))))((((.(((...))).)))).. ( -48.40) >DroGri_CAF1 73739 111 + 1 AAUGAUGCCAUUCAACUGACCCUUGGCCUCGCUGGCCAGUACGAGUGCCUCAAUGGCAUUGAUGUUGCCAUUGGUUUCCCGGAACGAGGCAUGCUCCGUAUCCGUGCGCAU ....(((((((............(((((.....)))))(((((.((((((((((((((.......))))))).((((....)))))))))))....)))))..))).)))) ( -41.00) >DroMoj_CAF1 89739 111 + 1 AAUGAUGCCAUUCAACUGGCCCUUGGCCUCGCUGGCCAGCGCCAGCGCCUCGAUCGCAUUGAUAUUGCCAUUGGCUUCCUUCACGGAGCCGUGCUCCGUGUCCGUGGCAUU ...((((((((......((((..((((..(((((((....)))))))..(((((...)))))....))))..)))).....((((((((...))))))))...)))))))) ( -47.90) >DroAna_CAF1 101938 111 + 1 AAUAAUGCCGUUCAUUUGACCUUUGGCCUCACUGGCCAGGGCCAAGGCUUCGAUGGCGUUGAUGUUGCCGUUCGGGUCCUUCACGGAAGCCCGCUCCGCGUCCGAGCGGGC .......((((....(((.((((.((((.....)))))))).)))((((..(((((((.......)))))))..))))....))))..((((((((.......)))))))) ( -47.30) >consensus AAUGAUGCCAUUCAUCUGACCCUUGGCCUCGCUGGCCAGGGCCAGCGCCUCGAUGGCAUUGAUGUUGCCAUUGGGAUCCUUCACGGAGGCGCGCUCCGCAUCCGUGCGCGC ...(((((............((((((((.....))))))))...((((((((((.(((.......))).)))((....)).....))))))).....)))))......... (-23.62 = -23.98 + 0.37)



| Location | 14,592,121 – 14,592,232 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -46.47 |
| Consensus MFE | -23.99 |
| Energy contribution | -24.55 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14592121 111 - 22224390 GCCUCCGCGGAUGCUGAAAGUGCCGCCGUGAAGGAUCCCAAUGGCAAUAUCAAUGCUAUCGAAGCGUUGGUCCUGGCCAGCGAGGCCAAGGGUCAGAUGAACGGGAUCAUC ..((.(((((..((.......))..))))).))((((((..((((...(((((((((.....)))))))))(((((((.....)))).))))))).......))))))... ( -43.50) >DroVir_CAF1 71837 111 - 1 UCGCGCUCGGACACGGAGCACGGCUCCAUACGAGAGCCCAAUGGCAACAUCAAUGCUGUCGAGGCGCUGGCCCUGGCCAGCGAGGCCAAGGGCCAGCUGAAUGGCAUCAUU ..(((((.......(((((...)))))...(((.(((...(((....)))....))).))).)))(((((((((((((.....)))).)))))))))......))...... ( -52.90) >DroPse_CAF1 78727 111 - 1 GCGCGUACCGAUGCGGAACGCGCCUCCAUCAAGGAUCCCAAUGGCAACAUCAACGCCAUCGAGGCUCUGGCCUUGGCCAGUGAGGCCAAGGGUCAAAUGAAUGGCAUCAUU ((((((.(((...))).))))))..((((((..(((((...((((....(((..(((.....))).(((((....)))))))).)))).)))))...)).))))....... ( -44.90) >DroGri_CAF1 73739 111 - 1 AUGCGCACGGAUACGGAGCAUGCCUCGUUCCGGGAAACCAAUGGCAACAUCAAUGCCAUUGAGGCACUCGUACUGGCCAGCGAGGCCAAGGGUCAGUUGAAUGGCAUCAUU ((((((.(((.((((.((..((((((.....((....))(((((((.......))))))))))))))))))))))))((((..((((...)))).))))....)))).... ( -43.00) >DroMoj_CAF1 89739 111 - 1 AAUGCCACGGACACGGAGCACGGCUCCGUGAAGGAAGCCAAUGGCAAUAUCAAUGCGAUCGAGGCGCUGGCGCUGGCCAGCGAGGCCAAGGGCCAGUUGAAUGGCAUCAUU .((((((....((((((((...))))))))......(((.((.(((.......))).))...)))((((((.((((((.....)))).)).))))))....)))))).... ( -49.50) >DroAna_CAF1 101938 111 - 1 GCCCGCUCGGACGCGGAGCGGGCUUCCGUGAAGGACCCGAACGGCAACAUCAACGCCAUCGAAGCCUUGGCCCUGGCCAGUGAGGCCAAAGGUCAAAUGAACGGCAUUAUU ((((((((.......))))))))..((((.((((...(((..(((.........))).)))...))))((((.(((((.....)))))..))))......))))....... ( -45.00) >consensus GCGCGCACGGACACGGAGCACGCCUCCGUGAAGGAACCCAAUGGCAACAUCAAUGCCAUCGAGGCGCUGGCCCUGGCCAGCGAGGCCAAGGGUCAGAUGAAUGGCAUCAUU ....((.((...........((((((......(....)..((((((.......)))))).))))))((((((((((((.....)))).)))))))).....))))...... (-23.99 = -24.55 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:09 2006