
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,583,635 – 14,583,813 |
| Length | 178 |
| Max. P | 0.991949 |

| Location | 14,583,635 – 14,583,735 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.38 |
| Mean single sequence MFE | -33.58 |
| Consensus MFE | -19.25 |
| Energy contribution | -21.33 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.991949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14583635 100 + 22224390 UGUACUUUGUA----AAUUUCGAUUAUUCCAAGAUCCAAUGUUGCU----------------GGAUGCUGGUCGUGUUGCUGGGAGCAGUCUGCAACAUUUGUUGCUGGCUCUCUCAGCG ...........----......(((....(((..(((((.......)----------------))))..)))....)))((((((((.((((.(((((....))))).)))))))))))). ( -35.00) >DroSec_CAF1 58949 96 + 1 ----CGUUAUA----AACUUCGAUUAUUCCAAAAUCCAAUGUUGCU----------------GGAUGCUGGUCGUGUUGCUGGGAGCAGUCUGCAACAUUUGUUGCUGGCUCUCUCAGCG ----.......----(((..(((.....(((..(((((.......)----------------))))..)))))).)))((((((((.((((.(((((....))))).)))))))))))). ( -36.50) >DroSim_CAF1 59002 96 + 1 ----CGUUAUA----AACUUCGAUUAUUCCAAGAUCCAAUGUUGCU----------------GGAUGCUGGUCGUGUUCCUGGGAGCAGUCUGCAACAUUUGUUGCUGGCUCUCUCAGCG ----.......----(((..(((.....(((..(((((.......)----------------))))..)))))).))).(((((((.((((.(((((....))))).))))))))))).. ( -31.10) >DroEre_CAF1 53823 100 + 1 AGUACAAUGCAAUGCAAUUGGAAUUAUUUCUAGGUCCAAUGUUGCU----------------GGAUGCUGUUCGUGUUGCUGGGAGCAGCCUGCAACAUUUGUUGCUAGCUCUCUC---- ........(((((((..((((((....))))))(((((.......)----------------)))).......))))))).(((((.(((..(((((....)))))..))))))))---- ( -32.80) >DroYak_CAF1 55627 114 + 1 UGUACAAUACAAUGCAUUUGCAAUUGUUUCUGGGUCCAAUGUUGCUGGAUGCUCAUGUUGCUGGAUGCUGUUCGUGUUGCUGGAAGCAGCCUGCAACAUUUGUAGCUGGCUCU--C---- ....((..(((((((....)).)))))...))...........((..(.(((..(((((((.((.((((.((((......)))))))).)).)))))))..))).)..))...--.---- ( -32.50) >consensus _GUACAUUAUA____AAUUUCGAUUAUUCCAAGAUCCAAUGUUGCU________________GGAUGCUGGUCGUGUUGCUGGGAGCAGUCUGCAACAUUUGUUGCUGGCUCUCUCAGCG ....................((((....(((..((((.........................))))..)))....))))(((((((.((((.(((((....))))).))))))))))).. (-19.25 = -21.33 + 2.08)

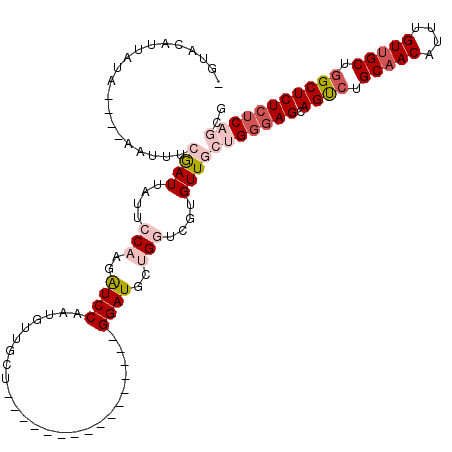
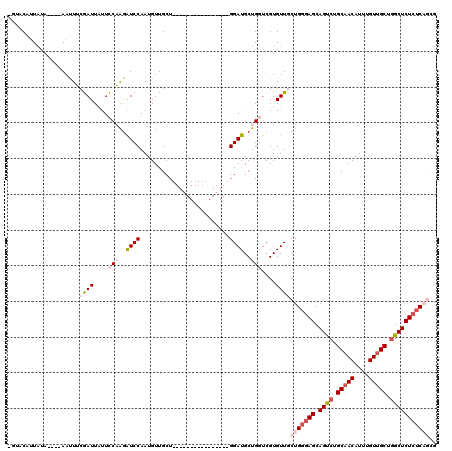
| Location | 14,583,635 – 14,583,735 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.38 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -10.94 |
| Energy contribution | -12.62 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14583635 100 - 22224390 CGCUGAGAGAGCCAGCAACAAAUGUUGCAGACUGCUCCCAGCAACACGACCAGCAUCC----------------AGCAACAUUGGAUCUUGGAAUAAUCGAAAUU----UACAAAGUACA .((((.(((((.(.(((((....))))).).)).))).))))....(((((((.((((----------------((.....)))))).)))).....))).....----........... ( -28.00) >DroSec_CAF1 58949 96 - 1 CGCUGAGAGAGCCAGCAACAAAUGUUGCAGACUGCUCCCAGCAACACGACCAGCAUCC----------------AGCAACAUUGGAUUUUGGAAUAAUCGAAGUU----UAUAACG---- .((((.(((((.(.(((((....))))).).)).))).))))....(((((((.((((----------------((.....)))))).)))).....))).....----.......---- ( -28.00) >DroSim_CAF1 59002 96 - 1 CGCUGAGAGAGCCAGCAACAAAUGUUGCAGACUGCUCCCAGGAACACGACCAGCAUCC----------------AGCAACAUUGGAUCUUGGAAUAAUCGAAGUU----UAUAACG---- ..(((.(((((.(.(((((....))))).).)).))).)))((((.(((((((.((((----------------((.....)))))).)))).....)))..)))----)......---- ( -25.20) >DroEre_CAF1 53823 100 - 1 ----GAGAGAGCUAGCAACAAAUGUUGCAGGCUGCUCCCAGCAACACGAACAGCAUCC----------------AGCAACAUUGGACCUAGAAAUAAUUCCAAUUGCAUUGCAUUGUACU ----(.(((((((.(((((....))))).)))).))).)......((((...(((...----------------.(((..((((((..((....))..)))))))))..))).))))... ( -28.30) >DroYak_CAF1 55627 114 - 1 ----G--AGAGCCAGCUACAAAUGUUGCAGGCUGCUUCCAGCAACACGAACAGCAUCCAGCAACAUGAGCAUCCAGCAACAUUGGACCCAGAAACAAUUGCAAAUGCAUUGUAUUGUACA ----.--.......(((....(((((((.((.((((....(.....)....)))).)).))))))).))).(((((.....))))).......(((((.((....)))))))........ ( -26.90) >consensus CGCUGAGAGAGCCAGCAACAAAUGUUGCAGACUGCUCCCAGCAACACGACCAGCAUCC________________AGCAACAUUGGAUCUUGGAAUAAUCGAAAUU____UAUAACGUAC_ ..((((((...((((((((....))))).((.((((....(.....)....)))))).........................))).))))))............................ (-10.94 = -12.62 + 1.68)

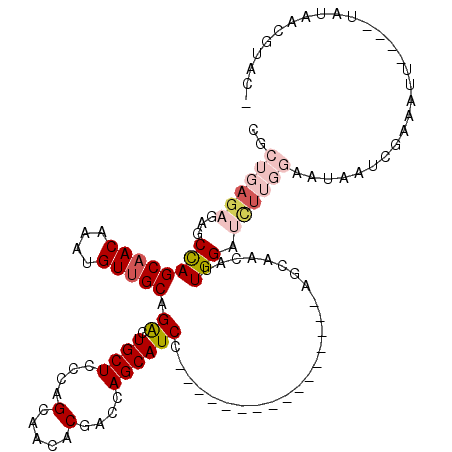

| Location | 14,583,671 – 14,583,775 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -22.58 |
| Energy contribution | -25.30 |
| Covariance contribution | 2.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14583671 104 + 22224390 GUUGCU----------------GGAUGCUGGUCGUGUUGCUGGGAGCAGUCUGCAACAUUUGUUGCUGGCUCUCUCAGCGCGACUGUCAUCAUGUUGCCUUCGAUUCUCAGAGUGUUGAU ((.((.----------------.((((.(((((((...((((((((.((((.(((((....))))).))))))))))))))))))).))))..)).))..(((((.(.....).))))). ( -41.50) >DroSec_CAF1 58981 104 + 1 GUUGCU----------------GGAUGCUGGUCGUGUUGCUGGGAGCAGUCUGCAACAUUUGUUGCUGGCUCUCUCAGCGAGACUGGCAUCAUGUUGCCUUCGAUUCUCAGGGUGUUGAU ......----------------.(((((..(((...((((((((((.((((.(((((....))))).)))))))))))))))))..))))).....(((((.(.....))))))...... ( -47.00) >DroSim_CAF1 59034 104 + 1 GUUGCU----------------GGAUGCUGGUCGUGUUCCUGGGAGCAGUCUGCAACAUUUGUUGCUGGCUCUCUCAGCGAGACUGGCAUCAUGUUGCUGUCGAUUCUCACUGUGUUGAU ((.((.----------------.(((((..(((....(((((((((.((((.(((((....))))).))))))))))).)))))..)))))..)).)).((((((.(.....).)))))) ( -43.10) >DroEre_CAF1 53863 96 + 1 GUUGCU----------------GGAUGCUGUUCGUGUUGCUGGGAGCAGCCUGCAACAUUUGUUGCUAGCUCUCUC--------UCUCAUCAUGUUGCCUUCGAUUUUCAAAGUGUUGAU .....(----------------(((.((....((((.((..(((((.(((..(((((....)))))..))))))))--------...)).))))..)).))))....((((....)))). ( -26.80) >DroYak_CAF1 55667 110 + 1 GUUGCUGGAUGCUCAUGUUGCUGGAUGCUGUUCGUGUUGCUGGAAGCAGCCUGCAACAUUUGUAGCUGGCUCU--C--------UCUCAUCAUGUUGCCUUCGAUUUUCAAUGUGUUGAU ...((..(.(((..(((((((.((.((((.((((......)))))))).)).)))))))..))).)..))...--.--------..(((.(((((((...........))))))).))). ( -32.60) >consensus GUUGCU________________GGAUGCUGGUCGUGUUGCUGGGAGCAGUCUGCAACAUUUGUUGCUGGCUCUCUCAGCG_GACUGUCAUCAUGUUGCCUUCGAUUCUCAAAGUGUUGAU .......................((((.(((((......(((((((.((((.(((((....))))).)))))))))))...))))).)))).........(((((.(.....).))))). (-22.58 = -25.30 + 2.72)

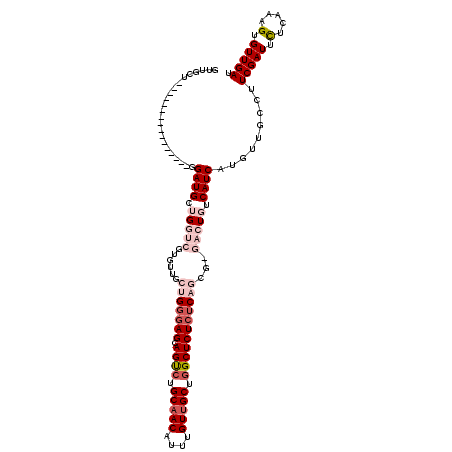
| Location | 14,583,695 – 14,583,813 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 89.59 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -26.82 |
| Energy contribution | -27.18 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14583695 118 + 22224390 UGGGAGCAGUCUGCAACAUUUGUUGCUGGCUCUCUCAGCGCGACUGUCAUCAUGUUGCCUUCGAUUCUCAGAGUGUUGAUAAGGGACUCAAUAAAUAUGUCAGAAAUAUGAGAUCUAU ((((((.((((.(((((....))))).))))))))))..(((((((....)).)))))....(((.(((.((((.((.....)).)))).....((((.......))))))))))... ( -34.70) >DroSec_CAF1 59005 118 + 1 UGGGAGCAGUCUGCAACAUUUGUUGCUGGCUCUCUCAGCGAGACUGGCAUCAUGUUGCCUUCGAUUCUCAGGGUGUUGAUAAGGGACUCAAUAAAUAUGUCAGAAAUAUGAGAUCUAU ((((((.((((.(((((....))))).))))))))))....((..((((......)))).))(((.((((.....((((((................)))))).....)))))))... ( -35.59) >DroSim_CAF1 59058 118 + 1 UGGGAGCAGUCUGCAACAUUUGUUGCUGGCUCUCUCAGCGAGACUGGCAUCAUGUUGCUGUCGAUUCUCACUGUGUUGAUAAGGGACUCAAUAAAUAUGUCAGAAAUAUGAGAUCUAU ((((((.((((.(((((....))))).))))))))))......(((((((..(((((..(((..((.(((......))).))..))).)))))...)))))))............... ( -35.40) >DroEre_CAF1 53887 110 + 1 UGGGAGCAGCCUGCAACAUUUGUUGCUAGCUCUCUC--------UCUCAUCAUGUUGCCUUCGAUUUUCAAAGUGUUGAUAAGGGACUCAAUAAAUAUGUCAGAAAUAUGAGAUCUAU .(((((.(((..(((((....)))))..))))))))--------..((.(((((((.((((((((.........)))))..)))(((...........)))...)))))))))..... ( -29.30) >DroYak_CAF1 55707 108 + 1 UGGAAGCAGCCUGCAACAUUUGUAGCUGGCUCU--C--------UCUCAUCAUGUUGCCUUCGAUUUUCAAUGUGUUGAUAAGGGACUCAAUAAAUAUGUCAAAAAUAUGAGAUCUAU .(..((.((((.((.((....)).)).))))))--.--------.)((.(((((((.((((((((.........)))))..)))(((...........)))...)))))))))..... ( -23.50) >consensus UGGGAGCAGUCUGCAACAUUUGUUGCUGGCUCUCUCAGCG_GACUGUCAUCAUGUUGCCUUCGAUUCUCAAAGUGUUGAUAAGGGACUCAAUAAAUAUGUCAGAAAUAUGAGAUCUAU .(((((.((((.(((((....))))).))))))))).............(((((((.((((((((.(.....).)))))..)))(((...........)))...)))))))....... (-26.82 = -27.18 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:05 2006