| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,617,523 – 1,617,615 |
| Length | 92 |
| Max. P | 0.884795 |

| Location | 1,617,523 – 1,617,615 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.05 |
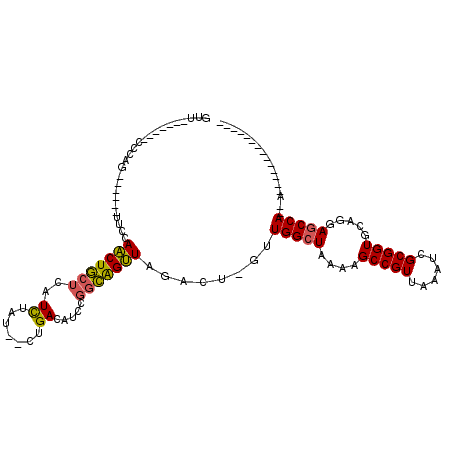
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -18.90 |
| Energy contribution | -18.57 |
| Covariance contribution | -0.33 |
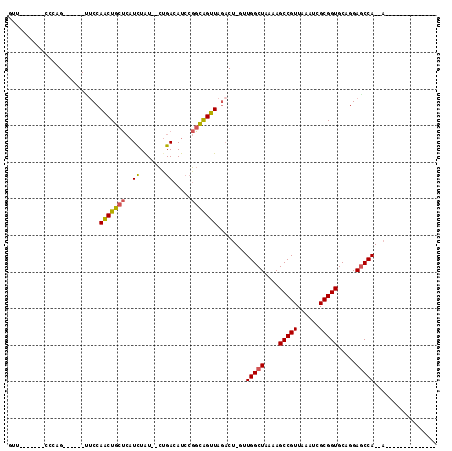
| Combinations/Pair | 1.21 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
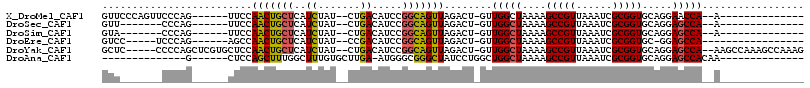
>X_DroMel_CAF1 1617523 92 + 22224390 GUUCCCAGUUCCCAG------UUCCAACUGCUCAUCUAU--CUGACAUCCGGCAGUUAGACU-GUUGGCUAAAAGCCGUUAAAUCGCGGUGCAGGAACCA--A-------------- (((((.((((..(((------((..(((((((..((...--..)).....))))))).))))-)..))))....(((((......)))))...)))))..--.-------------- ( -27.90) >DroSec_CAF1 13201 85 + 1 GUU-------CCCAG------UUCCAACUGCUCAUCUAU--CUGACAUCCGGCAGUUAGACU-GUUGGCUAAAAGCCGUUAAAUCGCGGUGCAGGAGCCA--A-------------- ...-------..(((------((..(((((((..((...--..)).....))))))).))))-)((((((....(((((......))))).....)))))--)-------------- ( -26.70) >DroSim_CAF1 29121 85 + 1 GUA-------CCCAG------UUCCAACUGCUCAUCUAU--CUGACAUCCGGCAGUUAGACU-GUUGGCUAAAAGCCGUUAAAUCGCGGUGCAGGAGCCA--A-------------- ...-------..(((------((..(((((((..((...--..)).....))))))).))))-)((((((....(((((......))))).....)))))--)-------------- ( -26.70) >DroEre_CAF1 13719 85 + 1 GUCC-----UCCCAG------AGCCAACUGCUCAUCUAU--CCGACAUCCGGCAGUUAGACU-GUUGGCUAAAAGCCGUUAAAUCGCGGUGC-GGAGCCA----------------- ...(-----((((..------(((((((..((...((..--(((.....))).))..))...-)))))))....(((((......)))))).-))))...----------------- ( -25.60) >DroYak_CAF1 15056 107 + 1 GCUC-----CCCCAGCUCGUGCUCCAACUGCUCAUCUAU--CUGACAUCCGGCAGUUAGACU-GUUGGCUAAAAGCCGUUAAAUCGCGGUGCAGGAGCCA--AAGCCAAAGCCAAAG (((.-----.....(((...(((((.(((((.......(--(((((........))))))..-...(((.....)))........)))))...)))))..--.)))...)))..... ( -29.10) >DroAna_CAF1 15279 82 + 1 --------------G------CUCCAGCUUUGGCUUUGUGCUUGA-AUGGGCGGGCUAUCCUGGCUGGCUAAAAGCCGUUAAAUCGCGGUGCAGGAGCCACAA-------------- --------------(------(((((((....)))...(((....-...(((.((((.....)))).)))....(((((......))))))))))))).....-------------- ( -29.00) >consensus GUU_______CCCAG______UUCCAACUGCUCAUCUAU__CUGACAUCCGGCAGUUAGACU_GUUGGCUAAAAGCCGUUAAAUCGCGGUGCAGGAGCCA__A______________ .........................(((((((..((.......)).....)))))))........(((((....(((((......))))).....)))))................. (-18.90 = -18.57 + -0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:51 2006