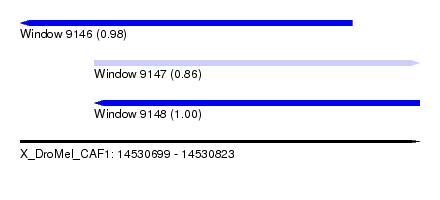
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,530,699 – 14,530,823 |
| Length | 124 |
| Max. P | 0.999939 |

| Location | 14,530,699 – 14,530,802 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.23 |
| Mean single sequence MFE | -25.81 |
| Consensus MFE | -22.94 |
| Energy contribution | -23.14 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14530699 103 - 22224390 AUUUAUGGGAACGUGCUUGGCUCAUUUCCAUUCGCUCACUUUCGGCAACAUUGUUCCCCGGAGAAUUCCACCAUUCUUCGGCAGAUACAAAAAAAAGAAACA----------------C- ......(((((((((..(((.......)))..)))........(....)...))))))((((((((......))))))))......................----------------.- ( -23.90) >DroSec_CAF1 5445 107 - 1 AUUUAUGGGAACGUGCUUGGCUCAUUUCCAUUCGCUCACUUUCGGCAACAUUGUUCCCCGGAGAAUUCCUCCAUUCUUCGGCAGAUACAAAAACCAUAUACACGUA------------C- ...((((((((((((..(((.......)))..)))........(....)...)))).(((((((((......)))))))))............)))))........------------.- ( -25.00) >DroSim_CAF1 5820 107 - 1 AUUUAUGGGAACGUGCUUGGCUCAUUUCCAUUCGCUCACUUUCGGCAACAUUGUUCCCCGGAGAAUUCCUCCAUUCUUCGGCAGAUACAAAAACCAUAUACACGUA------------C- ...((((((((((((..(((.......)))..)))........(....)...)))).(((((((((......)))))))))............)))))........------------.- ( -25.00) >DroEre_CAF1 5336 119 - 1 AUUUAUGGGAACGUGCUUGCCUCAUUUCCAUUCGCUCACUUUCGGCAACAUUGUUCCCCGGAGAAUUCCUCCAUUCUUCGGCAGAUACAAAAACG-UAUACACAUGCAGACGCAGAUACA ......(((((((((.(((((......................))))))).)))))))((((((((......))))))))..............(-(((.....(((....))).)))). ( -28.95) >DroYak_CAF1 5565 107 - 1 AUUUAUGGGAACGUGCUUGGCUCAUUUCCAUUCGCUGACUUUCGGCAACAUUGUUCCCCGGAGAAUUCCUCCAUUCUUCGGCAGAUACAAUAACC-UAUACAGAUA------------CA ...((((((...(((..(((.......)))..)((((.....)))).))(((((((.(((((((((......)))))))))..)).)))))..))-))))......------------.. ( -26.20) >consensus AUUUAUGGGAACGUGCUUGGCUCAUUUCCAUUCGCUCACUUUCGGCAACAUUGUUCCCCGGAGAAUUCCUCCAUUCUUCGGCAGAUACAAAAACCAUAUACACAUA____________C_ ......(((((((((..(((.......)))..)))........(....)...))))))((((((((......))))))))........................................ (-22.94 = -23.14 + 0.20)

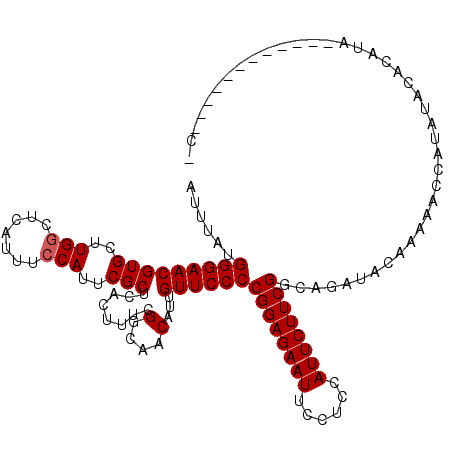
| Location | 14,530,722 – 14,530,823 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 97.62 |
| Mean single sequence MFE | -22.96 |
| Consensus MFE | -20.64 |
| Energy contribution | -20.84 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14530722 101 + 22224390 CGAAGAAUGGUGGAAUUCUCCGGGGAACAAUGUUGCCGAAAGUGAGCGAAUGGAAAUGAGCCAAGCACGUUCCCAUAAAUCAUAAAGGAAAGAAUUUACGA ((.(((((......))))).))((((((...(((..(....)..)))(..(((.......)))..)..))))))........................... ( -22.00) >DroSec_CAF1 5472 101 + 1 CGAAGAAUGGAGGAAUUCUCCGGGGAACAAUGUUGCCGAAAGUGAGCGAAUGGAAAUGAGCCAAGCACGUUCCCAUAAAUCAUAAAGAAAAGAAUUUACGA ((.....(((((.....)))))((((((...(((..(....)..)))(..(((.......)))..)..))))))........................)). ( -22.80) >DroSim_CAF1 5847 101 + 1 CGAAGAAUGGAGGAAUUCUCCGGGGAACAAUGUUGCCGAAAGUGAGCGAAUGGAAAUGAGCCAAGCACGUUCCCAUAAAUCAUAAAGAAAAGAAUUUACGA ((.....(((((.....)))))((((((...(((..(....)..)))(..(((.......)))..)..))))))........................)). ( -22.80) >DroEre_CAF1 5375 100 + 1 CGAAGAAUGGAGGAAUUCUCCGGGGAACAAUGUUGCCGAAAGUGAGCGAAUGGAAAUGAGGCAAGCACGUUCCCAUAAAUCAUAAAGAA-GGAAUUUACGA ((.....(((((.....)))))((((((..(((((((....((...(.....)..))..))).)))).))))))...............-........)). ( -21.90) >DroYak_CAF1 5592 101 + 1 CGAAGAAUGGAGGAAUUCUCCGGGGAACAAUGUUGCCGAAAGUCAGCGAAUGGAAAUGAGCCAAGCACGUUCCCAUAAAUCAUAAAGAAAAGAAUUUACGA ((.....(((((.....)))))((((((...((((.(....).))))(..(((.......)))..)..))))))........................)). ( -25.30) >consensus CGAAGAAUGGAGGAAUUCUCCGGGGAACAAUGUUGCCGAAAGUGAGCGAAUGGAAAUGAGCCAAGCACGUUCCCAUAAAUCAUAAAGAAAAGAAUUUACGA ((.(((((......))))).))((((((...(((..(....)..)))(..(((.......)))..)..))))))........................... (-20.64 = -20.84 + 0.20)


| Location | 14,530,722 – 14,530,823 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 97.62 |
| Mean single sequence MFE | -25.77 |
| Consensus MFE | -24.74 |
| Energy contribution | -24.94 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.69 |
| SVM RNA-class probability | 0.999939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14530722 101 - 22224390 UCGUAAAUUCUUUCCUUUAUGAUUUAUGGGAACGUGCUUGGCUCAUUUCCAUUCGCUCACUUUCGGCAACAUUGUUCCCCGGAGAAUUCCACCAUUCUUCG (((((((........))))))).....(((((((((..(((.......)))..)))........(....)...))))))((((((((......)))))))) ( -25.70) >DroSec_CAF1 5472 101 - 1 UCGUAAAUUCUUUUCUUUAUGAUUUAUGGGAACGUGCUUGGCUCAUUUCCAUUCGCUCACUUUCGGCAACAUUGUUCCCCGGAGAAUUCCUCCAUUCUUCG (((((((........))))))).....(((((((((..(((.......)))..)))........(....)...))))))((((((((......)))))))) ( -25.70) >DroSim_CAF1 5847 101 - 1 UCGUAAAUUCUUUUCUUUAUGAUUUAUGGGAACGUGCUUGGCUCAUUUCCAUUCGCUCACUUUCGGCAACAUUGUUCCCCGGAGAAUUCCUCCAUUCUUCG (((((((........))))))).....(((((((((..(((.......)))..)))........(....)...))))))((((((((......)))))))) ( -25.70) >DroEre_CAF1 5375 100 - 1 UCGUAAAUUCC-UUCUUUAUGAUUUAUGGGAACGUGCUUGCCUCAUUUCCAUUCGCUCACUUUCGGCAACAUUGUUCCCCGGAGAAUUCCUCCAUUCUUCG (((((((....-...))))))).....(((((((((.(((((......................))))))).)))))))((((((((......)))))))) ( -25.65) >DroYak_CAF1 5592 101 - 1 UCGUAAAUUCUUUUCUUUAUGAUUUAUGGGAACGUGCUUGGCUCAUUUCCAUUCGCUGACUUUCGGCAACAUUGUUCCCCGGAGAAUUCCUCCAUUCUUCG (((((((........))))))).....(((((((.(..(((.......)))..)((((.....)))).....)))))))((((((((......)))))))) ( -26.10) >consensus UCGUAAAUUCUUUUCUUUAUGAUUUAUGGGAACGUGCUUGGCUCAUUUCCAUUCGCUCACUUUCGGCAACAUUGUUCCCCGGAGAAUUCCUCCAUUCUUCG (((((((........))))))).....(((((((((..(((.......)))..)))........(....)...))))))((((((((......)))))))) (-24.74 = -24.94 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:45 2006