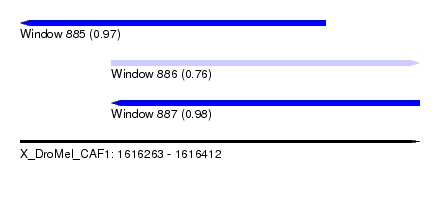
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,616,263 – 1,616,412 |
| Length | 149 |
| Max. P | 0.979439 |

| Location | 1,616,263 – 1,616,377 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.84 |
| Mean single sequence MFE | -39.55 |
| Consensus MFE | -21.44 |
| Energy contribution | -24.82 |
| Covariance contribution | 3.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1616263 114 - 22224390 GCAGGACUGGCGAUCAAGCUGUGUUGGCCAGCAAUCACAGCGGACCUGAAAAGGUCCUUGCCAAGGAGUCCAAGUGUUGAGUAUGUUCAUGUGUGAGUAUCUUAAAGGACUACU------ .......((((((....(((((((((.....))).))))))((((((....))))))))))))((.(((((.....(((((.(((((((....)))))))))))).))))).))------ ( -42.60) >DroSec_CAF1 11975 118 - 1 GCAGGACUGGCGAUCAAGCUGUGUUGGCCAGCAAUCACAGCGGACCUGAAAAGGUCCUUGCCAAGGAGUCCAAGUGUUG--CAUGUCCAUGUGCGAGUAUCUUAAAGGAGGACUAAUCUC ...(((.((((((....(((((((((.....))).))))))((((((....))))))))))))...(((((.....(((--(((......))))))...(((....))))))))..))). ( -44.10) >DroSim_CAF1 27884 118 - 1 GCAGGACUGGCGAUCAAGCUGUGUUGGCCAGCAAUCACAGCGGACCUGAAAAGGUCCUUGCCAAGGAGUCCAAGUGUUG--CAUGUCCAUGUGCGAGUAUCUUAAAGGACGACUACUCUC ...((((((((((....(((((((((.....))).))))))((((((....)))))))))))....))))).......(--(((......))))((((((((....)))....))))).. ( -44.20) >DroYak_CAF1 13799 98 - 1 GCAGGACUGGCGAUCAAGCUGUUG---------------GGGCCCAGCA----AUCACAGCAAAGGAGCCCAAGUAGCCAG-GCAUCCAUG--UACAUAUCUUAAAGGGUGAAUACCUAC ..(((.(((((......(((....---------------((((((.((.----......))...)).)))).))).)))))-.(((((...--.............)))))....))).. ( -27.29) >consensus GCAGGACUGGCGAUCAAGCUGUGUUGGCCAGCAAUCACAGCGGACCUGAAAAGGUCCUUGCCAAGGAGUCCAAGUGUUG__CAUGUCCAUGUGCGAGUAUCUUAAAGGACGACUACUCUC ...((((((((......(((((((((.....))).))))))((((((....))))))..)))....)))))............(((((..................)))))......... (-21.44 = -24.82 + 3.38)

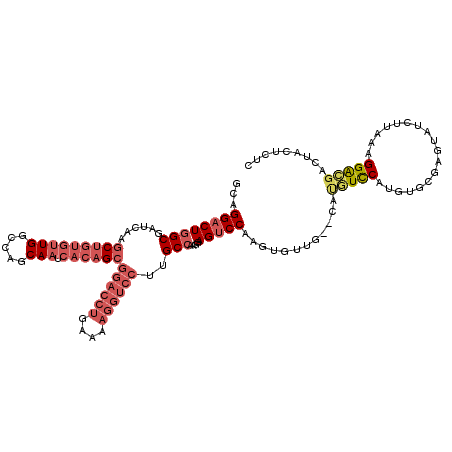
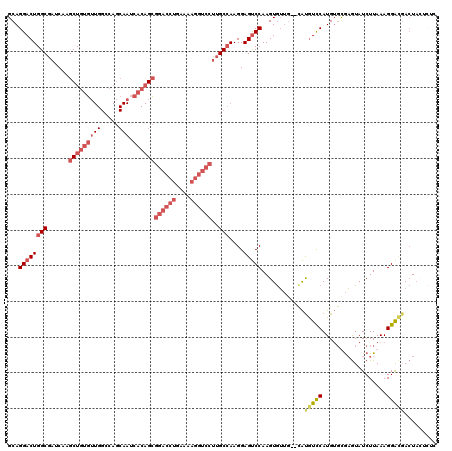
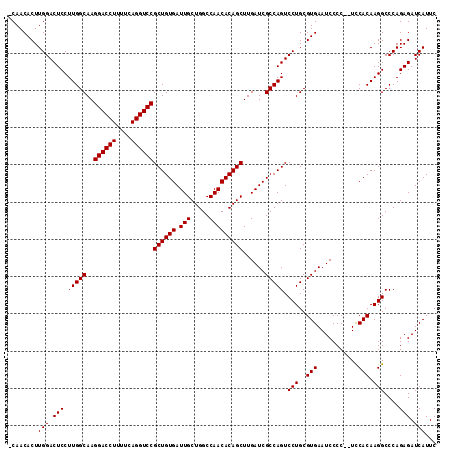
| Location | 1,616,297 – 1,616,412 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 97.66 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -34.20 |
| Energy contribution | -34.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1616297 115 + 22224390 UCAACACUUGGACUCCUUGGCAAGGACCUUUUCAGGUCCGCUGUGAUUGCUGGCCAACACAGCUUGAUCGCCAGUCCUGCGUGAAUCCCCCGUCCACAAGGCCCAGAGAUCAUUC ....(((..(((((....(((..((((((....))))))((((((.(((.....)))))))))......))))))))...)))......((........)).............. ( -34.50) >DroSec_CAF1 12014 112 + 1 -CAACACUUGGACUCCUUGGCAAGGACCUUUUCAGGUCCGCUGUGAUUGCUGGCCAACACAGCUUGAUCGCCAGUCCUGCGUGAAUCCCC--UCCACAAGGCCCAGAGAUCAUUC -...(((..(((((....(((..((((((....))))))((((((.(((.....)))))))))......))))))))...))).....((--(.....))).............. ( -34.60) >DroSim_CAF1 27923 112 + 1 -CAACACUUGGACUCCUUGGCAAGGACCUUUUCAGGUCCGCUGUGAUUGCUGGCCAACACAGCUUGAUCGCCAGUCCUGCGUGAAUCCCC--UUCACAAGGCCCAGAGAUCAUUC -.......(((.(((.(((((..((((((....))))))((((((.(((.....)))))))))......))))).(((..(((((.....--))))).)))....))).)))... ( -37.10) >consensus _CAACACUUGGACUCCUUGGCAAGGACCUUUUCAGGUCCGCUGUGAUUGCUGGCCAACACAGCUUGAUCGCCAGUCCUGCGUGAAUCCCC__UCCACAAGGCCCAGAGAUCAUUC ........(((.(((.(((((..((((((....))))))((((((.(((.....)))))))))......))))).(((..(((...........))).)))....))).)))... (-34.20 = -34.20 + -0.00)

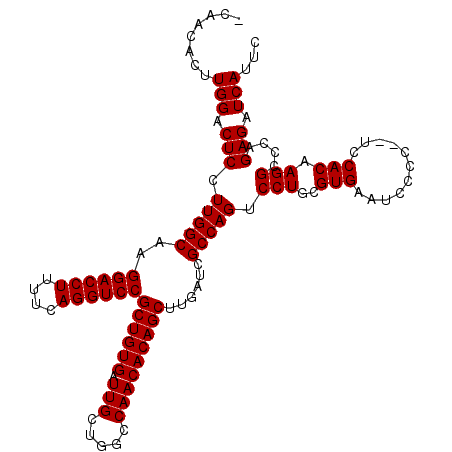
| Location | 1,616,297 – 1,616,412 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 97.66 |
| Mean single sequence MFE | -46.50 |
| Consensus MFE | -44.57 |
| Energy contribution | -44.90 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
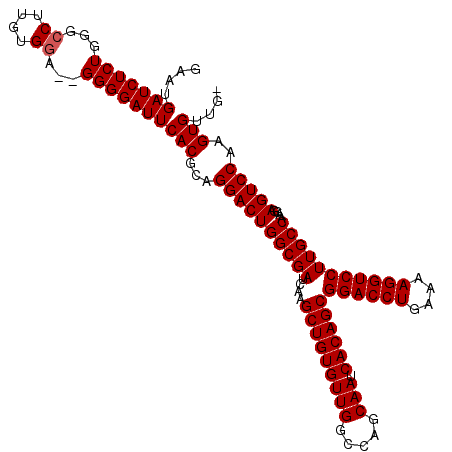
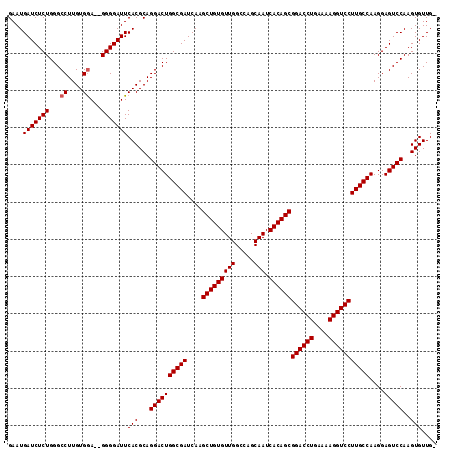
>X_DroMel_CAF1 1616297 115 - 22224390 GAAUGAUCUCUGGGCCUUGUGGACGGGGGAUUCACGCAGGACUGGCGAUCAAGCUGUGUUGGCCAGCAAUCACAGCGGACCUGAAAAGGUCCUUGCCAAGGAGUCCAAGUGUUGA ....(((((((.(.((....)).).)))))))(((...((((((((((....(((((((((.....))).))))))((((((....)))))))))))....)))))..))).... ( -48.40) >DroSec_CAF1 12014 112 - 1 GAAUGAUCUCUGGGCCUUGUGGA--GGGGAUUCACGCAGGACUGGCGAUCAAGCUGUGUUGGCCAGCAAUCACAGCGGACCUGAAAAGGUCCUUGCCAAGGAGUCCAAGUGUUG- ....(((((((...((....)).--)))))))(((...((((((((((....(((((((((.....))).))))))((((((....)))))))))))....)))))..)))...- ( -46.60) >DroSim_CAF1 27923 112 - 1 GAAUGAUCUCUGGGCCUUGUGAA--GGGGAUUCACGCAGGACUGGCGAUCAAGCUGUGUUGGCCAGCAAUCACAGCGGACCUGAAAAGGUCCUUGCCAAGGAGUCCAAGUGUUG- ...((..((((.(((((((((((--.....)))))).))).))(((((....(((((((((.....))).))))))((((((....)))))))))))..))))..)).......- ( -44.50) >consensus GAAUGAUCUCUGGGCCUUGUGGA__GGGGAUUCACGCAGGACUGGCGAUCAAGCUGUGUUGGCCAGCAAUCACAGCGGACCUGAAAAGGUCCUUGCCAAGGAGUCCAAGUGUUG_ ....(((((((...((....))...)))))))(((...((((((((((....(((((((((.....))).))))))((((((....)))))))))))....)))))..))).... (-44.57 = -44.90 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:50 2006