
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,508,235 – 14,508,329 |
| Length | 94 |
| Max. P | 0.663457 |

| Location | 14,508,235 – 14,508,329 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 68.35 |
| Mean single sequence MFE | -29.63 |
| Consensus MFE | -13.35 |
| Energy contribution | -14.72 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
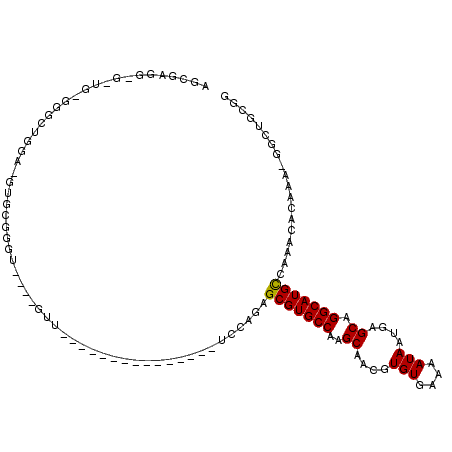
>X_DroMel_CAF1 14508235 94 + 22224390 GGGGAUGUG-UG-GGGGUGGAGGUGCGGGU----UUU---------------UCCAGAGCGUGCCAAGCUACGUGUGAAAAUAAUGAGCAGGCAUGCCAAACACAAA-GGCUGCGG (.((...((-((-....((((((.......----.))---------------))))(.(((((((..(((...(((....)))...))).))))))))...))))..-..)).).. ( -30.10) >DroVir_CAF1 22540 85 + 1 GGCG-------G-GAGCAGGC-GGGCCGG-----GAG---------------CCUAGAGCGUGCCAAGCAACGUGUGAAAAUAAUGAGCGGGCAUGCUCAACACAAA-GGCCGUG- .((.-------.-..)).(((-((((...-----..)---------------))).(((((((((..((....(((....)))....)).)))))))))........-.)))...- ( -34.50) >DroGri_CAF1 29875 89 + 1 AGCG-------G---GUAGGU-GUACCGAGAGGGGUA---------------GCCGGAGCGUGCCAAGCAACGUGUGAAAAUAAUGAGCAGGCAUGCUUAACACAAUAGACGGGG- ..((-------.---.(((((-...((....))....---------------))).(((((((((..((....(((....)))....)).))))))))).......))..))...- ( -28.00) >DroEre_CAF1 29796 92 + 1 A-UGGGGGAUGG-GGGAUG------GGGAAUGGGGUU---------------UCCAGAGCGUGCCAAGCUACGUGUGAAAAUAAUGAGCAGGCAUGCCAAACACAAA-GGCUGCGG .-((.((..(..-((..((------.((((......)---------------)))...(((((((..(((...(((....)))...))).)))))))))..).)..)-..)).)). ( -24.70) >DroYak_CAF1 37552 99 + 1 AGUGAGGGGCUG-GGGCUGGGGGUGGGGGUGGAGUUU---------------UCCAGAGCGUGCCAAGCUACGUGUGAAAAUAAUGAGCAGGCAUGCCAAACACAAA-GGCUGCGG .........(((-.((((....(((....(((.....---------------.)))(.(((((((..(((...(((....)))...))).))))))))...)))...-)))).))) ( -33.20) >DroAna_CAF1 39382 110 + 1 GUGAAGGUG-UUAGGGUUGGAGUUGGGGGC----GUUACUAAAAGGGGCGGGGCCAGAGCGUGCCAAGCAACGUGUGAAAAUAAUGAGCAGGCAUGUCAAACACAAA-GGCCGCGA .....(((.-((.(..(((........(((----.(..((......))..).)))...(((((((..((....(((....)))....)).))))))))))...).))-.))).... ( -27.30) >consensus AGCGAGG_G_UG_GGGCUGGA_GUGCGGGU____GUU_______________UCCAGAGCGUGCCAAGCAACGUGUGAAAAUAAUGAGCAGGCAUGCCAAACACAAA_GGCUGCGG ..........................................................(((((((..((....(((....)))....)).)))))))................... (-13.35 = -14.72 + 1.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:34 2006