| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,498,997 – 14,499,108 |
| Length | 111 |
| Max. P | 0.615576 |

| Location | 14,498,997 – 14,499,108 |
|---|---|
| Length | 111 |
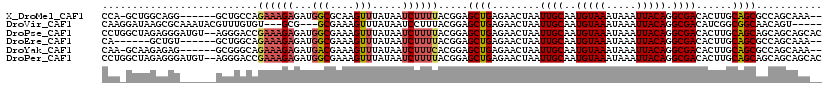
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.41 |
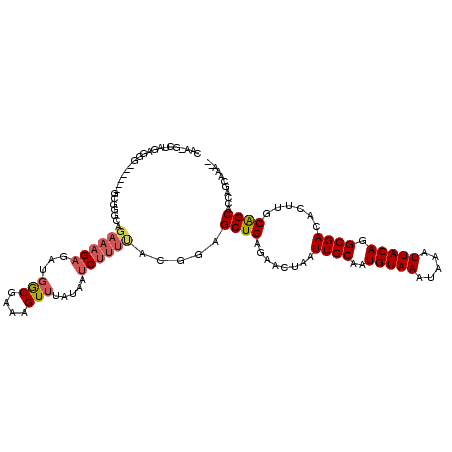
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -16.71 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14498997 111 + 22224390 CCA-GCUGGCAGG------GCUGCCAGAAAGAGAUGGCGCAAGUUUAUAAUCUUUUACGGAGCUGAGAACUAAUUGCAAUGUAAAUAAAUUACAGGCGACACUUGCAGCGCCAGCAAA-- ...-((((((..(------(((.((.((((((...(((....))).....))))))..)))))).........(((((((((((.....)))))(....)..)))))).))))))...-- ( -35.00) >DroVir_CAF1 12732 109 + 1 CAAGGAUAAGCGCAAAUACGUUUGUGU---GCG---GCGAAAGUUUAUAAUCCUUUACGGAGCUGAGAACUAAUUGCAAUGUAAAUAAAUUACAGGCGACAUCGGCGGCAACAGU----- .((((((..(((((((....)))))))---..(---((....)))....))))))......(((((.......((((..(((((.....))))).))))..)))))(....)...----- ( -26.00) >DroPse_CAF1 25405 118 + 1 CCUGGCUAGAGGGAUGU--AGGGACCGAAAGAGAUGGCGAAAGUUUAUAAUCUUUUACGGAGCUGAGAACUAAUUGCAAUGUAAAUAAAUUACAGGCGACACUUGCAGCAGCAGCAGCAC (((.((.........))--.))).(((.((((((((((....)))....))))))).))).((((........((((..(((((.....))))).))))...((((....)))))))).. ( -28.90) >DroEre_CAF1 21305 106 + 1 CA------GCUGU------GCUGGCAGAAAGAGAUGGCGAAAGUUUAUAAUCUUUUACGGAGCUGAGAACUAAUUGCAAUGUAAAUAAAUUACAGGCGACACUUGCAGCGCCAGCAAA-- ..------....(------((((((.((((((...(((....))).....)))))).....((((........((((..(((((.....))))).))))......)))))))))))..-- ( -32.94) >DroYak_CAF1 28091 111 + 1 CAA-GCAAGAGAG------GCGGGCAGAAAGAGAUGACGAAAGUUUAUAAUCUUUCACGGAGCUGAGAACUAAUUGCAAUGUAAAUAAAUUACAGGCGACACUUGCAGCGCCAGCAAA-- ...-.........------((.(((.((((((...(((....))).....)))))).....((((........((((..(((((.....))))).))))......))))))).))...-- ( -30.44) >DroPer_CAF1 25579 118 + 1 CCUGGCUAGAGGGAUGU--AGGGACCGAAAGAGAUGGCGAAAGUUUAUAAUCUUUUACGGAGCUGAGAACUAAUUGCAAUGUAAAUAAAUUACAGGCGACACUUGCAGCAGCAGCAGCAC (((.((.........))--.))).(((.((((((((((....)))....))))))).))).((((........((((..(((((.....))))).))))...((((....)))))))).. ( -28.90) >consensus CAA_GCUAGAGGG______GCGGGCAGAAAGAGAUGGCGAAAGUUUAUAAUCUUUUACGGAGCUGAGAACUAAUUGCAAUGUAAAUAAAUUACAGGCGACACUUGCAGCACCAGCAAA__ ..........................((((((...(((....))).....)))))).....((((........((((..(((((.....))))).))))......))))........... (-16.71 = -17.32 + 0.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:31 2006