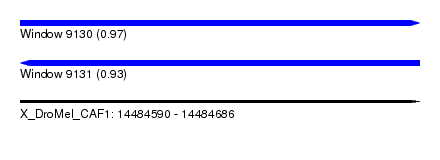
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,484,590 – 14,484,686 |
| Length | 96 |
| Max. P | 0.971226 |

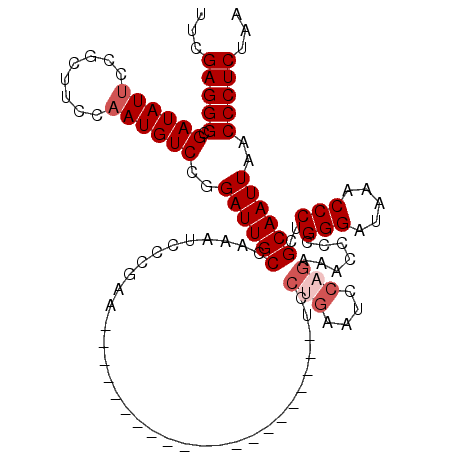
| Location | 14,484,590 – 14,484,686 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -27.40 |
| Energy contribution | -30.28 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
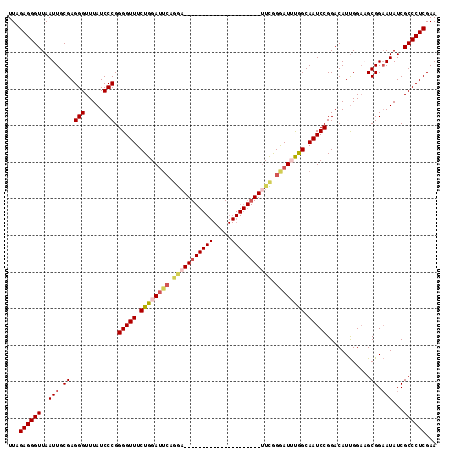
>X_DroMel_CAF1 14484590 96 + 22224390 UUAGAGGGUUAAUUGCGAGGGUUUAUCCCGGGGUUUCUGGAUUCUGGA---------------------UUCGGGAUUUGGCAAUCCGGACAUUGGACGCGGAAUAUCGCCCUCGAA ...((((((...(((((.(((.....))).(((((.(..(((((((..---------------------..)))))))..).)))))..........)))))......))))))... ( -34.80) >DroSec_CAF1 9944 89 + 1 UUAGAGGGUUAAUUGCGAGGGUUUAUCCCGGGGUUUCUGGA----------------------------UUCGGGAUAUAGCAAUCCGGACAUUGGAAGCGGAAUAUCGCCCUCGAA ...((((((..(((.(..(((.....)))...(((((..(.----------------------------.((.((((......)))).))..)..)))))).)))...))))))... ( -30.80) >DroSim_CAF1 16434 96 + 1 UUAGAGGGUUAAUUGCGAGGGUUUAUCCCGGGGUUUCUGGAUUCAGGA---------------------UUCGGGAUAUAGCAAUCCGGACAUUGGAAGCGGAAUAUCGCCCUCGAA ...((((((..(((.((...((((((((((((..(((((....)))))---------------------))))))))).)))..((((.....))))..)).)))...))))))... ( -32.50) >DroEre_CAF1 8275 96 + 1 UUAGAGGGUUAAUUGCGAGGGUUUAUCCCGGGGUUUCUGGAUUCAGAA---------------------CUUAGAAUUCGGCAAUCCGGACAUCGGACGCGGCAUAUCGCCCUCGAA ...((((((....(((..(((.....))).(((((.((((((((....---------------------....)))))))).)))))..............)))....))))))... ( -33.80) >DroYak_CAF1 13280 117 + 1 UUAGAGGGUUAAUUGCGAGGGUUUAUCCCGGGGUUUCUGGAUUCAGGAUUCUGAAUUCAGGAUUCAAGAUUUGGAAUUCGGCAAUCCGGACAUUGGUAGCGGAAUAUCGCCCUCGAA ...((((((...((((..(((.....)))..(..(((..((((..((((((((....))))))))..))))..)))..).))))((((.((....))..)))).....))))))... ( -42.10) >consensus UUAGAGGGUUAAUUGCGAGGGUUUAUCCCGGGGUUUCUGGAUUCAGGA_____________________UUCGGGAUUUGGCAAUCCGGACAUUGGAAGCGGAAUAUCGCCCUCGAA ...((((((..(((.((.(((.....))).(((((.((((((((.(((((((((((.....))))))))))).)))))))).)))))............)).)))...))))))... (-27.40 = -30.28 + 2.88)

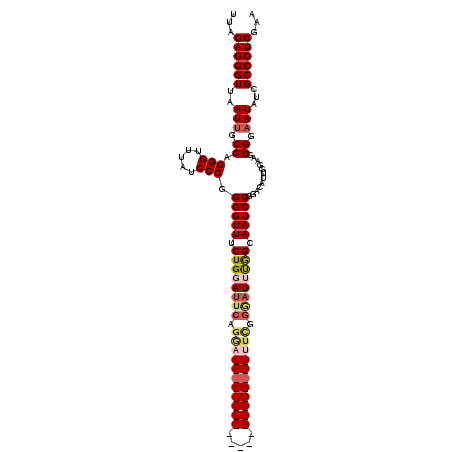
| Location | 14,484,590 – 14,484,686 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -18.86 |
| Energy contribution | -19.86 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14484590 96 - 22224390 UUCGAGGGCGAUAUUCCGCGUCCAAUGUCCGGAUUGCCAAAUCCCGAA---------------------UCCAGAAUCCAGAAACCCCGGGAUAAACCCUCGCAAUUAACCCUCUAA ..((((((...(((((((..((.....((.(((((...........))---------------------))).)).....)).....)))))))..))))))............... ( -22.70) >DroSec_CAF1 9944 89 - 1 UUCGAGGGCGAUAUUCCGCUUCCAAUGUCCGGAUUGCUAUAUCCCGAA----------------------------UCCAGAAACCCCGGGAUAAACCCUCGCAAUUAACCCUCUAA ..((((((.........((.(((.......)))..))..(((((((..----------------------------...........)))))))..))))))............... ( -23.62) >DroSim_CAF1 16434 96 - 1 UUCGAGGGCGAUAUUCCGCUUCCAAUGUCCGGAUUGCUAUAUCCCGAA---------------------UCCUGAAUCCAGAAACCCCGGGAUAAACCCUCGCAAUUAACCCUCUAA ..((((((.........((.(((.......)))..))..(((((((..---------------------..(((....)))......)))))))..))))))............... ( -25.50) >DroEre_CAF1 8275 96 - 1 UUCGAGGGCGAUAUGCCGCGUCCGAUGUCCGGAUUGCCGAAUUCUAAG---------------------UUCUGAAUCCAGAAACCCCGGGAUAAACCCUCGCAAUUAACCCUCUAA ...(((((.....(((.(((((((.....)))).))).((........---------------------(((((....))))).....(((.....)))))))).....)))))... ( -26.60) >DroYak_CAF1 13280 117 - 1 UUCGAGGGCGAUAUUCCGCUACCAAUGUCCGGAUUGCCGAAUUCCAAAUCUUGAAUCCUGAAUUCAGAAUCCUGAAUCCAGAAACCCCGGGAUAAACCCUCGCAAUUAACCCUCUAA ..((((((...(((((((.........((.(((((...........))))).))...(((.((((((....)))))).)))......)))))))..))))))............... ( -30.10) >consensus UUCGAGGGCGAUAUUCCGCUUCCAAUGUCCGGAUUGCCAAAUCCCGAA_____________________UCCUGAAUCCAGAAACCCCGGGAUAAACCCUCGCAAUUAACCCUCUAA ...(((((.((((((........))))))..((((((..................................(((....))).......(((.....)))..))))))..)))))... (-18.86 = -19.86 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:29 2006