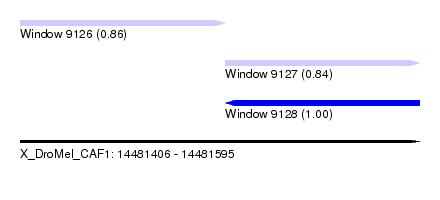
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,481,406 – 14,481,595 |
| Length | 189 |
| Max. P | 0.996271 |

| Location | 14,481,406 – 14,481,503 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 84.69 |
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -23.04 |
| Energy contribution | -23.04 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14481406 97 + 22224390 ACAAUUAAAAUGACAACGGAGUAGAUGGCGAGGCGAAAGGAUUUUUGCACGUUUCGCUUUCUACUCAUUGCGCCCC--------AUGUCCC----CUCGCC-CAAUCCAU ..................(((((((.(((((((((..............))))))))).)))))))((((.((...--------.......----...)).-)))).... ( -24.06) >DroSec_CAF1 6756 106 + 1 ACAAUUAAAAUGACAACGGAGUAGAUGGCGAGGCGAAAGGAUUUUUGCACGUUUCGCUUUCUACUCAUGGCGCCCUCGUCCUCAUUGUCCC----CUCCCCACAAUCCAU ...........(((((.((((((((.(((((((((..............))))))))).)))))))..((((....))))..).)))))..----............... ( -29.24) >DroSim_CAF1 13210 106 + 1 ACAAUUAAAAUGACAACGGAGUAGAUGGCGAGGCGAAAGGAUUUUUGCACGUUUCGCUUUCUACUCAUGGCGCCCUCGUCCUCAUUGUCCC----CUCCCCGCAAUCCAU ...........(((((.((((((((.(((((((((..............))))))))).)))))))..((((....))))..).)))))..----............... ( -29.24) >DroEre_CAF1 5108 83 + 1 ACAAUUAAAAUGACAACGGAGUAGAUGGCGAGGCGAAAGGAUUUUUGCACGUUUCGCUUUCUACUCAUUG---------------------------CCACACAAUCCAU ..........((.(((..(((((((.(((((((((..............))))))))).))))))).)))---------------------------.)).......... ( -23.94) >DroYak_CAF1 10042 105 + 1 ACAAUUAAAAUGACAACGGAGUAGAUGGCGAGGCGAAAGGAUUUUUGCACGUUUCGCUUUCUACUCAUUGGGACCCCG----C-AUGUCCCCCUCCUCCGCACAAUCCAU ................(((((((((.(((((((((..............))))))))).)))))))...(((((....----.-..))))).......)).......... ( -30.24) >consensus ACAAUUAAAAUGACAACGGAGUAGAUGGCGAGGCGAAAGGAUUUUUGCACGUUUCGCUUUCUACUCAUUGCGCCCCCG____C_AUGUCCC____CUCCCCACAAUCCAU ..................(((((((.(((((((((..............))))))))).)))))))............................................ (-23.04 = -23.04 + -0.00)

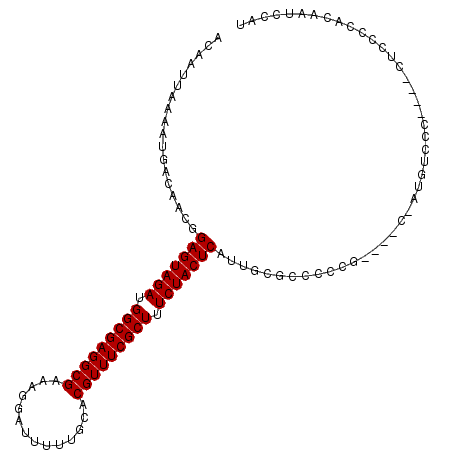

| Location | 14,481,503 – 14,481,595 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -18.03 |
| Consensus MFE | -13.28 |
| Energy contribution | -13.79 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14481503 92 + 22224390 UCAUCCUUAUGCCAGUGA--AAGAAAAAAUACACGAG----------UGAAAAAAAAAUGGGAA-AAAUUCAUGAAAUGUUACUGAUGAAAACUGACAUAAGGCG ....(((((((.((((..--...........((..((----------(((.......(((((..-...)))))......)))))..))...)))).))))))).. ( -15.41) >DroSec_CAF1 6862 91 + 1 UCAUCCUUAUGCCAGUGG--AAGGUAAAAUCCACGAG----------UGAAAA-AAAAUGGGAA-AAAUUCAUGAAAUGUUACUGAUGAAAACUGACAUAAGGCG ....(((((((.((((((--(........))))).((----------(((...-...(((((..-...)))))......))))).........)).))))))).. ( -20.10) >DroSim_CAF1 13316 91 + 1 UCAUCCUUAUGCCAGUGG--AAGAAAAAAUCCACGAG----------UGAAAA-AAAAUGGGAA-AAAUUCAUGAAAUGUUACUGAUGAAAACUGACAUAAGGCG ....(((((((.((((((--(........))))).((----------(((...-...(((((..-...)))))......))))).........)).))))))).. ( -20.10) >DroYak_CAF1 10147 103 + 1 UCAUCCUUAUCCCAGUGGGGAAGGAAAAGUCCACGAGUGCAAAAAAA-AAAAA-AAAAUGGGAAAAAAUUCAUGAAAUGUUACUGAUGAAAACUGACAUAAGGCG ....(((((((((((((((..........)))))(....).......-.....-....))))).....(((((.(........).)))))........))))).. ( -16.50) >consensus UCAUCCUUAUGCCAGUGG__AAGAAAAAAUCCACGAG__________UGAAAA_AAAAUGGGAA_AAAUUCAUGAAAUGUUACUGAUGAAAACUGACAUAAGGCG ....(((((((.((((.............((((.........................))))......(((((.(........).))))).)))).))))))).. (-13.28 = -13.79 + 0.50)


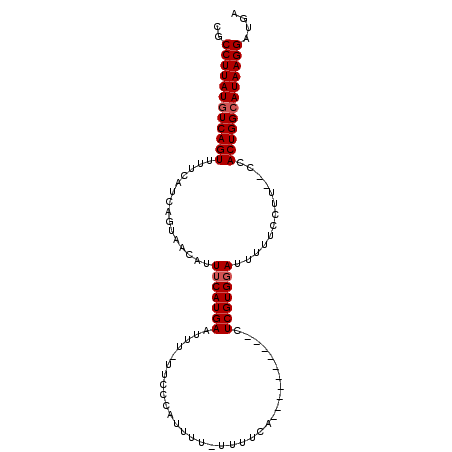
| Location | 14,481,503 – 14,481,595 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -19.98 |
| Consensus MFE | -17.48 |
| Energy contribution | -17.98 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14481503 92 - 22224390 CGCCUUAUGUCAGUUUUCAUCAGUAACAUUUCAUGAAUUU-UUCCCAUUUUUUUUUCA----------CUCGUGUAUUUUUUCUU--UCACUGGCAUAAGGAUGA ..((((((((((((.(((((.((......)).)))))...-................(----------(....))..........--..)))))))))))).... ( -18.30) >DroSec_CAF1 6862 91 - 1 CGCCUUAUGUCAGUUUUCAUCAGUAACAUUUCAUGAAUUU-UUCCCAUUUU-UUUUCA----------CUCGUGGAUUUUACCUU--CCACUGGCAUAAGGAUGA ..((((((((((...(((((.((......)).)))))...-..........-......----------...(((((........)--)))))))))))))).... ( -22.40) >DroSim_CAF1 13316 91 - 1 CGCCUUAUGUCAGUUUUCAUCAGUAACAUUUCAUGAAUUU-UUCCCAUUUU-UUUUCA----------CUCGUGGAUUUUUUCUU--CCACUGGCAUAAGGAUGA ..((((((((((...(((((.((......)).)))))...-..........-......----------...(((((........)--)))))))))))))).... ( -22.40) >DroYak_CAF1 10147 103 - 1 CGCCUUAUGUCAGUUUUCAUCAGUAACAUUUCAUGAAUUUUUUCCCAUUUU-UUUUU-UUUUUUUGCACUCGUGGACUUUUCCUUCCCCACUGGGAUAAGGAUGA ..((((((.(((((.(((((.((......)).)))))..............-.....-.............(.(((........))).)))))).)))))).... ( -16.80) >consensus CGCCUUAUGUCAGUUUUCAUCAGUAACAUUUCAUGAAUUU_UUCCCAUUUU_UUUUCA__________CUCGUGGAUUUUUCCUU__CCACUGGCAUAAGGAUGA ..((((((((((((...............(((((((.................................))))))).............)))))))))))).... (-17.48 = -17.98 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:27 2006