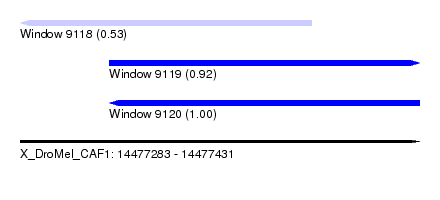
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,477,283 – 14,477,431 |
| Length | 148 |
| Max. P | 0.998720 |

| Location | 14,477,283 – 14,477,391 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 87.64 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -22.52 |
| Energy contribution | -23.36 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14477283 108 - 22224390 UGUGGAUGACGCCUCUCAGUCGGCGUCAGGUGCAUCAUAGGUGUGAAGUGCAAUCAAAACCUACACUCGGAAAAC-----UCAAAGUUCGCCCUAAAAUUUAUAUUUAAA--CCA (((((.(((((((........))))))).(..(.((((....)))).)..).........)))))...((..(((-----.....)))..))..................--... ( -26.00) >DroSec_CAF1 2499 107 - 1 UGUGGAUGACGCCUCCCAGUAGGCGUCAGGUGCAUCAUAGGUGUGAAGUGCAAUCAA-ACCUGCACUCGGAAAAC-----UCAAAGUUCGCCCUAAAAUUUAUAUUUACA--AUA ((((((((((((((......)))))))..........((((.((((((((((.....-...)))))...((....-----))....)))))))))........)))))))--... ( -31.40) >DroSim_CAF1 8997 108 - 1 UGUGGAUGACGCCUCUCAGUCGGCGUCAGGUGCAUCAUAGGUGUGAAGUGCAAUCAAAACCUGCACUCGGAAAAC-----UCAAAGUUCGCCCUAAAAUUUAUAUUUACA--AUA (((((((((((((........))))))..........((((.((((((((((.........)))))...((....-----))....)))))))))........)))))))--... ( -29.70) >DroEre_CAF1 130 113 - 1 UGUGGAUGACGCCUCUCAGUCGGCGUCAGGUGCAUCAUAGGUGUGAAGUGCAAUCAAAAGCUACACUCAGAAAACUUCACUCGAACUUCUCCCCAAAAGUUAUAGCUUGG--GCA ((((..(((((((........)))))))..))))......(.(((((((........................))))))).).........((((..(((....))))))--).. ( -28.26) >DroYak_CAF1 5145 110 - 1 CGUGAAUGACGCCUCUCAGUCGGCGUCAGGUGCAUCAUAGGUGUGAAGUGCAAUCAAAACCUGCACUCGGAAAAC-----UCGAACUUCUCCCUGCAAGUUAUAUUUAAAAAAAA .(((..(((((((........)))))))..)))....((((.(.((((((((.........)))..((((.....-----))))))))).))))).................... ( -28.40) >consensus UGUGGAUGACGCCUCUCAGUCGGCGUCAGGUGCAUCAUAGGUGUGAAGUGCAAUCAAAACCUGCACUCGGAAAAC_____UCAAAGUUCGCCCUAAAAUUUAUAUUUAAA__ACA .(((..(((((((........)))))))..)))....((((.((((((((((.........)))))...((.........))....))))))))).................... (-22.52 = -23.36 + 0.84)



| Location | 14,477,316 – 14,477,431 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 94.11 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -26.58 |
| Energy contribution | -26.02 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.924928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14477316 115 + 22224390 GUUUUCCGAGUGUAGGUUUUGAUUGCACUUCACACCUAUGAUGCACCUGACGCCGACUGAGAGGCGUCAUCCACAUCGUUUUCUUCUUUUUUUUUGCCACUUUGCGCAACUUCUU .......((((((((.......)))))))).......((((((....(((((((........)))))))....))))))..............(((((.....).))))...... ( -26.60) >DroSec_CAF1 2532 109 + 1 GUUUUCCGAGUGCAGGU-UUGAUUGCACUUCACACCUAUGAUGCACCUGACGCCUACUGGGAGGCGUCAUCCACAUCAUUUUCU-----UUUUUGGCCACUUUGCGCAACUUCUU .........((((((((-.(((.......))).))))((((((....((((((((......))))))))....)))))).....-----..............))))........ ( -31.10) >DroSim_CAF1 9030 110 + 1 GUUUUCCGAGUGCAGGUUUUGAUUGCACUUCACACCUAUGAUGCACCUGACGCCGACUGAGAGGCGUCAUCCACAUCGUUUUCU-----UUUUUGGCCACUUUGCGCAACUUCUU .........((((((((..(((.......))).))))((((((....(((((((........)))))))....)))))).....-----..............))))........ ( -28.00) >DroEre_CAF1 168 109 + 1 GUUUUCUGAGUGUAGCUUUUGAUUGCACUUCACACCUAUGAUGCACCUGACGCCGACUGAGAGGCGUCAUCCACAUCGUUUUCU-----U-UUUGGCCACUUUGCGCAACUUCUU .......(((((((((....).)))))))).......((((((....(((((((........)))))))....)))))).....-----.-.(((((......)).)))...... ( -25.80) >DroYak_CAF1 5180 110 + 1 GUUUUCCGAGUGCAGGUUUUGAUUGCACUUCACACCUAUGAUGCACCUGACGCCGACUGAGAGGCGUCAUUCACGUCGUUUUCU-----UUUUUGGCCACUUUGCGCAACUUCUU .........((((((((..(((.......))).))))((((((....(((((((........)))))))....)))))).....-----..............))))........ ( -27.60) >consensus GUUUUCCGAGUGCAGGUUUUGAUUGCACUUCACACCUAUGAUGCACCUGACGCCGACUGAGAGGCGUCAUCCACAUCGUUUUCU_____UUUUUGGCCACUUUGCGCAACUUCUU .......((((((((.......)))))))).......((((((....(((((((........)))))))....))))))................(((.....).))........ (-26.58 = -26.02 + -0.56)


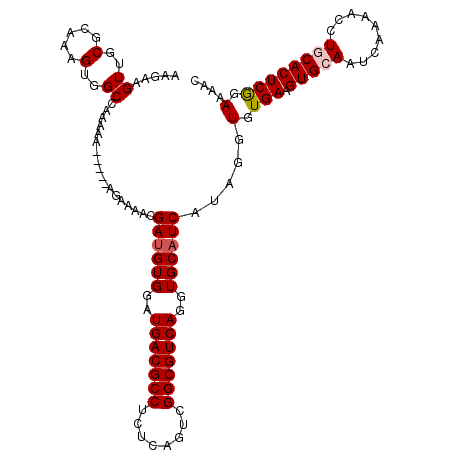
| Location | 14,477,316 – 14,477,431 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 94.11 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -28.64 |
| Energy contribution | -29.08 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14477316 115 - 22224390 AAGAAGUUGCGCAAAGUGGCAAAAAAAAAGAAGAAAACGAUGUGGAUGACGCCUCUCAGUCGGCGUCAGGUGCAUCAUAGGUGUGAAGUGCAAUCAAAACCUACACUCGGAAAAC .....((..(.....)..)).................((((((((.(((((((........))))))).(..(.((((....)))).)..).........))))).)))...... ( -31.50) >DroSec_CAF1 2532 109 - 1 AAGAAGUUGCGCAAAGUGGCCAAAAA-----AGAAAAUGAUGUGGAUGACGCCUCCCAGUAGGCGUCAGGUGCAUCAUAGGUGUGAAGUGCAAUCAA-ACCUGCACUCGGAAAAC .....((..(.....)..))......-----.....((((((((..((((((((......))))))))..))))))))...(.(((.(((((.....-...)))))))).).... ( -34.90) >DroSim_CAF1 9030 110 - 1 AAGAAGUUGCGCAAAGUGGCCAAAAA-----AGAAAACGAUGUGGAUGACGCCUCUCAGUCGGCGUCAGGUGCAUCAUAGGUGUGAAGUGCAAUCAAAACCUGCACUCGGAAAAC .....((..(.....)..))......-----.......((((((..(((((((........)))))))..)))))).....(.(((.(((((.........)))))))).).... ( -31.50) >DroEre_CAF1 168 109 - 1 AAGAAGUUGCGCAAAGUGGCCAAA-A-----AGAAAACGAUGUGGAUGACGCCUCUCAGUCGGCGUCAGGUGCAUCAUAGGUGUGAAGUGCAAUCAAAAGCUACACUCAGAAAAC .....((..(.....)..))....-.-----.......((((((..(((((((........)))))))..))))))...((((((....((........))))))))........ ( -29.30) >DroYak_CAF1 5180 110 - 1 AAGAAGUUGCGCAAAGUGGCCAAAAA-----AGAAAACGACGUGAAUGACGCCUCUCAGUCGGCGUCAGGUGCAUCAUAGGUGUGAAGUGCAAUCAAAACCUGCACUCGGAAAAC .....((..(.....)..))......-----......(((.(((..(((((((........))))))).(..(.((((....)))).)..)............))))))...... ( -28.40) >consensus AAGAAGUUGCGCAAAGUGGCCAAAAA_____AGAAAACGAUGUGGAUGACGCCUCUCAGUCGGCGUCAGGUGCAUCAUAGGUGUGAAGUGCAAUCAAAACCUGCACUCGGAAAAC .....((..(.....)..))..................((((((..(((((((........)))))))..)))))).....(.(((.(((((.........)))))))).).... (-28.64 = -29.08 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:20 2006