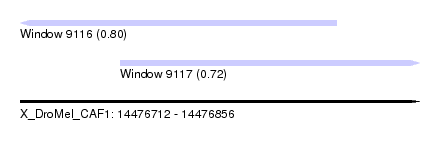
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,476,712 – 14,476,856 |
| Length | 144 |
| Max. P | 0.795717 |

| Location | 14,476,712 – 14,476,826 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -23.99 |
| Consensus MFE | -22.03 |
| Energy contribution | -22.28 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
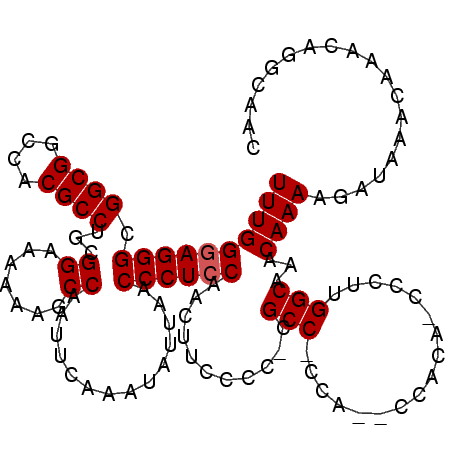
>X_DroMel_CAF1 14476712 114 - 22224390 UUUGGGAGGGCGGCGGCCACGCCUGCGGAAAAAAGCCAAUUCAAAUAUUAACCCUCCAACUUCCCC-CGCC-CCACCCCACA-CCCUUGGCAAACAAAAGAUAAACAAACAGGCAAU ((((((((((.((((....)))).((........))...............)))))).........-.(((-..........-.....)))...))))................... ( -24.56) >DroSec_CAF1 1950 112 - 1 UUUGGGAGGGCGGCGGCCACGCCUGCGGAAAAAACCCAAUUCAAAUAUUAACCCUCCAACUUCCCC-CGCC-CCA--CCACA-CCCUUGGCAAACAAAAGAUAAACAAACAGGCAAC ((((((((((.((((....))))((.((......)))).............)))))).........-.(((-...--.....-.....)))...))))................... ( -24.92) >DroSim_CAF1 8448 113 - 1 UUUGGGAGGGCGGCGGCCACGCCUGCGGAAAAAAGCCAAUUCAAAUAUUAACCCUCCAACUUCUCC-CGCCCCCA--CCACU-CCCUUGGCAAACAAAAGAUAAACAAACAGGCAAC ....((((((.((((....)))).((........))...............)))))).........-.(((....--....(-(..(((.....)))..))..........)))... ( -24.87) >DroYak_CAF1 4585 112 - 1 UUUGGAAGGGCGGCGGCCACGCCUGUGGAAAAAAGCCAAUUCAAAUAUUAACCCUCCAACUUCCCCCCGCC-C----CCACACCCCUUGGCAAACAAAAGAUAAACAAACAAGCAAC .(((((.(((.((((....))))..(((.......))).............)))))))).........(((-.----...........))).......................... ( -21.60) >consensus UUUGGGAGGGCGGCGGCCACGCCUGCGGAAAAAAGCCAAUUCAAAUAUUAACCCUCCAACUUCCCC_CGCC_CCA__CCACA_CCCUUGGCAAACAAAAGAUAAACAAACAGGCAAC ((((((((((.((((....))))...((.......))..............))))))...........(((.................)))...))))................... (-22.03 = -22.28 + 0.25)


| Location | 14,476,748 – 14,476,856 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.40 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -20.88 |
| Energy contribution | -21.62 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14476748 108 + 22224390 UGGGGUGG-GGCG-GGGGAAGUUGGAGGGUUAAUAUUUGAAUUGGCUUUUUUCCGCAGGCGUGGCCGCCGCCCUCCCAAAAAAAAAAUAAGUAU-AAA---------AAAAUAACGACGA ..(((.((-((((-(.((.....(((((((((((......))))))))))).((((....)))))).)))))))))).................-...---------............. ( -35.00) >DroSec_CAF1 1986 102 + 1 UGG--UGG-GGCG-GGGGAAGUUGGAGGGUUAAUAUUUGAAUUGGGUUUUUUCCGCAGGCGUGGCCGCCGCCCUCCCAAAAAAAA-----GUAUUAAA---------AAAAUACCGACGA (((--..(-((((-(.((.....(((((.(((((......))))).))))).((((....)))))).))))))..))).......-----(((((...---------..)))))...... ( -31.40) >DroSim_CAF1 8484 104 + 1 UGG--UGGGGGCG-GGAGAAGUUGGAGGGUUAAUAUUUGAAUUGGCUUUUUUCCGCAGGCGUGGCCGCCGCCCUCCCAAAAAAAA----UGUAUAAAA---------AAAAUAACGACGA (((--.(((((((-(.....(..(((((((((((......)))))))))))..)...(((...))).))))))))))).......----.........---------............. ( -32.30) >DroYak_CAF1 4622 108 + 1 UGG----G-GGCGGGGGGAAGUUGGAGGGUUAAUAUUUGAAUUGGCUUUUUUCCACAGGCGUGGCCGCCGCCCUUCCAAAAA-------UGUAUAAAAAAAAAAUAGAAAAUAACGACGA (((----(-(((((.((......(((((((((((......))))))))))).((((....)))))).))))))..)))....-------............................... ( -30.40) >consensus UGG__UGG_GGCG_GGGGAAGUUGGAGGGUUAAUAUUUGAAUUGGCUUUUUUCCGCAGGCGUGGCCGCCGCCCUCCCAAAAAAAA____UGUAUAAAA_________AAAAUAACGACGA (((...((.(((..(.((((....((((((((((......)))))))))))))).).((((....))))))))).))).......................................... (-20.88 = -21.62 + 0.75)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:18 2006