| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,470,300 – 14,470,407 |
| Length | 107 |
| Max. P | 0.666078 |

| Location | 14,470,300 – 14,470,407 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -21.96 |
| Consensus MFE | -18.01 |
| Energy contribution | -18.57 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14470300 107 - 22224390 UUAUGCUUGGGGGGUGUAUGGGGUAUUUUUGCUUUAAUUAAAUUUUACACAAAUAUUUUGUGCGAUGCGAAAGUAUU-UCCCAAAUUGAAGUUCUAUGCCGAC-GU-----------AGC ......(((((..((((((..(((((((.((................)).)))))))..))))...((....)))).-.))))).........(((((....)-))-----------)). ( -24.19) >DroPse_CAF1 326 104 - 1 UUAUUUU----G---------GGCAUUUUUACUUUAAUUAAAUUUUACACAAAUAUUUUCUG---UGCAAAAGUAUUUUCCCAAAUUGAAGUUCUAUGCCGUUCAGUCCAUGGCAAUAUC (((.(((----(---------((......((((((............((((.........))---))..))))))....)))))).))).......((((((.......))))))..... ( -16.14) >DroSec_CAF1 944 103 - 1 UUAUGUU----GGGUGUAUGGGGUAUUUUUGCUUUAAUUAAAUUUUACACAAAUAUUUUGUGCGAUGCGAAAGUAUU-UCCCAAAUUGAAGUUCUAUGCCGAC-GU-----------AGC (((((((----((...((((((........(((((((((........(((((.....))))).(((((....)))))-.....))))))))))))))))))))-))-----------)). ( -25.10) >DroSim_CAF1 7440 103 - 1 UUAUGUU----GGGUGUAUGGGGUAUUUUUGCUUUAAUUAAAUUUUACACAAAUAUUUUGUGCGAUGCGAAAGUAUU-UCCCAAAUUGAAGUUCUAUGCCGAC-GU-----------AGC (((((((----((...((((((........(((((((((........(((((.....))))).(((((....)))))-.....))))))))))))))))))))-))-----------)). ( -25.10) >DroYak_CAF1 3554 107 - 1 UUAUGUU----GGGUGUAUGGGGUAUUUUUGCUUUAAUUAAAUUUUACACAAAUAUUUUGUGCGAUGCGAAAGUAUU-UCCCAAAUUGAAGUUCUAUGCCGAC-GUUCUA-------UGC .......----(..(((....(((((....(((((((((........(((((.....))))).(((((....)))))-.....)))))))))...))))).))-)..)..-------... ( -25.10) >DroPer_CAF1 326 104 - 1 UUAUUUU----G---------GGCAUUUUUACUUUAAUUAAAUUUUACACAAAUAUUUUCUG---UGCAAAAGUAUUUUCCCAAAUUGAAGUUCUAUGCCGUUCAGUCCAUGGCAAUAUC (((.(((----(---------((......((((((............((((.........))---))..))))))....)))))).))).......((((((.......))))))..... ( -16.14) >consensus UUAUGUU____GGGUGUAUGGGGUAUUUUUGCUUUAAUUAAAUUUUACACAAAUAUUUUGUGCGAUGCGAAAGUAUU_UCCCAAAUUGAAGUUCUAUGCCGAC_GU___________AGC .....................(((((....(((((((((........(((((.....))))).(((((....)))))......)))))))))...))))).................... (-18.01 = -18.57 + 0.56)


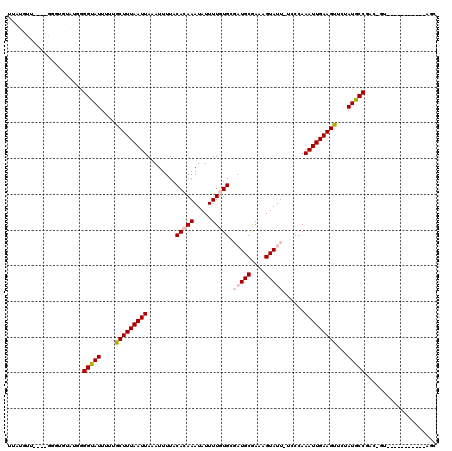
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:15 2006