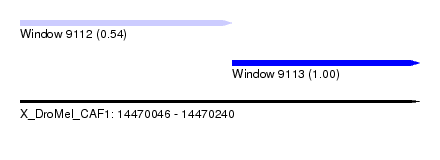
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,470,046 – 14,470,240 |
| Length | 194 |
| Max. P | 0.997813 |

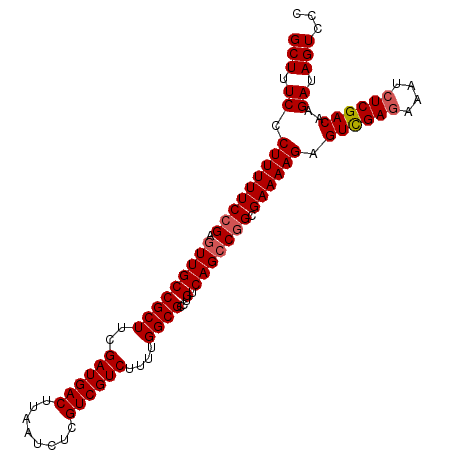
| Location | 14,470,046 – 14,470,149 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -28.89 |
| Energy contribution | -29.45 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14470046 103 + 22224390 GCUUUCCCUUUUUCCGAGUUGCCGCUUCGAUGACUUAAUCUCGUCGUCUUUUGGCGCCUGUCAGCCGGCGAAAAGAGUUGAGAAAUCUCGACAAGAUAGUCCC (((.((.(((((((((.(((((((((..((((((........))))))....))))...).))))))).)))))).((((((....))))))..)).)))... ( -31.80) >DroSec_CAF1 693 103 + 1 GCUUUCCCUUUUUCCGAGUUGCCGCUUCGAUGACUUAAUCUCGUCGUCUUUUGGCGCCUGUCAGCCGGCGAAAAGAGUCGAAAAAUCUCGACAAGAUAGUCCC (((.((.(((((((((.(((((((((..((((((........))))))....))))...).))))))).)))))).(((((......)))))..)).)))... ( -30.40) >DroSim_CAF1 7194 103 + 1 GCUUUCCCUUUUUCCGAGUUGCCGCUUCGAUGACUUAAUCUCGUCGUCUUUUGGCGCCUGUCAGCCGGCGAAAAGAGUCGAGAAAUCUCGACAAGAUAGUCCC (((.((.(((((((((.(((((((((..((((((........))))))....))))...).))))))).)))))).((((((....))))))..)).)))... ( -33.90) >DroYak_CAF1 3307 103 + 1 GCUUUCCCUUUUUCCGAGUUGCCGCUUCGAUGACUUAAUCUCGUCGUCUUUUGGCGCCUGUCAGGCUGCGAAAAGAGUCGAGAAAUCUCGACAAGAUAGUCCC (((.((.((((((.((....((((....((((((........))))))...))))(((.....))))).)))))).((((((....))))))..)).)))... ( -31.30) >consensus GCUUUCCCUUUUUCCGAGUUGCCGCUUCGAUGACUUAAUCUCGUCGUCUUUUGGCGCCUGUCAGCCGGCGAAAAGAGUCGAGAAAUCUCGACAAGAUAGUCCC (((.((.(((((((((.(((((((((..((((((........))))))....))))...).))))))).)))))).((((((....))))))..)).)))... (-28.89 = -29.45 + 0.56)

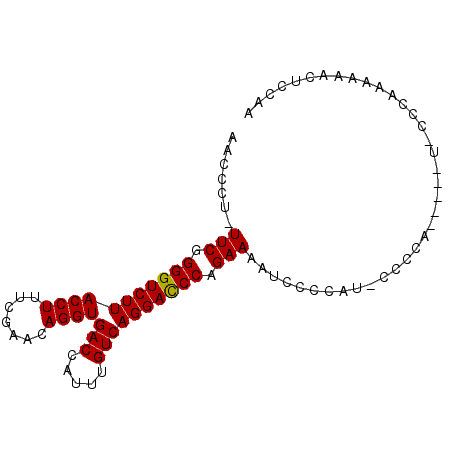
| Location | 14,470,149 – 14,470,240 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 84.76 |
| Mean single sequence MFE | -17.02 |
| Consensus MFE | -16.29 |
| Energy contribution | -16.10 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.94 |
| SVM RNA-class probability | 0.997813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14470149 91 + 22224390 AACCCU-UUCGGGGUCUUACCUUUCGAACAGGUGACCAUUUGUCAGGACCCAGAAAAUUCCCAUUCCCCAUCCCAAAAACAAAAAACUCCAA .....(-(((.(((((((((((.......))))(((.....)))))))))).)))).................................... ( -17.70) >DroSec_CAF1 796 89 + 1 AACCCU-UUCGGGGUCUUACCUUUCGAACAGGUGACCAUUUGUCAGGACCCAGAAAAUCCCCAU-CCCCAUCCCAU-CCCAAAAAACUCCAA .....(-(((.(((((((((((.......))))(((.....)))))))))).))))........-...........-............... ( -17.70) >DroSim_CAF1 7297 84 + 1 AACCCU-UUCGGGGUCUUACCUUUCGAACAGGUGACCAUUUGUCAGGACCCAGAAAAUCCCCAU-CCCCA-----U-CCCAAAAAACUCCAA .....(-(((.(((((((((((.......))))(((.....)))))))))).))))........-.....-----.-............... ( -17.70) >DroYak_CAF1 3410 78 + 1 AACCCUUUUCGGGGUCUUACCUUUCGAACAGGUGACCAUUUGUCAGGAUCCAGAAAUC-----U-UCCCA-----U-CCCAAA--GCAUCAA ......((((.(((((((((((.......))))(((.....)))))))))).))))..-----.-.....-----.-......--....... ( -15.00) >consensus AACCCU_UUCGGGGUCUUACCUUUCGAACAGGUGACCAUUUGUCAGGACCCAGAAAAUCCCCAU_CCCCA_____U_CCCAAAAAACUCCAA .......(((.(((((((((((.......))))(((.....)))))))))).)))..................................... (-16.29 = -16.10 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:14 2006