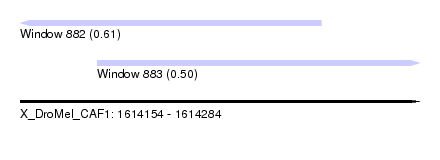
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,614,154 – 1,614,284 |
| Length | 130 |
| Max. P | 0.608505 |

| Location | 1,614,154 – 1,614,252 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 85.89 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -19.46 |
| Energy contribution | -20.39 |
| Covariance contribution | 0.93 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
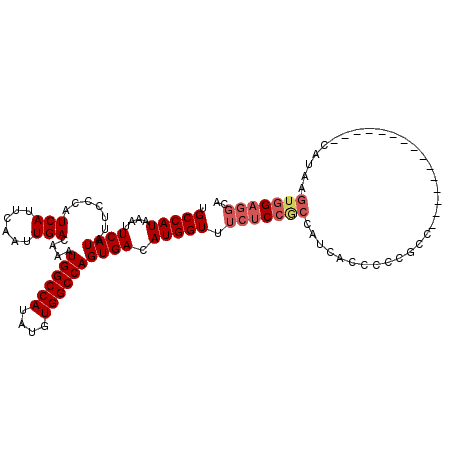
>X_DroMel_CAF1 1614154 98 - 22224390 UGCCAUAAAUUCAUAUUCCCAUCAUUCAAUUGACAAAUGGCCAUAUGUGGCCAGUGACAUGGUUUCUCCGCCAUCACCCCCGCC---------------CAUAAGUGGAGGCA .(((.................(((......)))....((((((....))))))((((..((((......))))))))..((((.---------------.....)))).))). ( -25.90) >DroSec_CAF1 9832 98 - 1 UGCCAUAAAUUCAUAUUCCCAUCAUUCAAUUGACAAAUGGCCAUAUGUGGCCAGUGACAUGGUUUCUCCGCCAUCACCCCCGCC---------------CAUAAGUGGAGGCA .(((.................(((......)))....((((((....))))))((((..((((......))))))))..((((.---------------.....)))).))). ( -25.90) >DroSim_CAF1 25792 98 - 1 UGCCAUAAAUUCAUAUUCCCAUCAUUCAAUUGACAAAUGGCCAUAUGUGGCCAGUGACAUGGUUUCUCCGCCAUAACCCCCGCC---------------CAUAAGUGGAGGCA .(((......((((.......(((......)))....((((((....)))))))))).(((((......))))).....((((.---------------.....)))).))). ( -26.00) >DroEre_CAF1 10573 98 - 1 UGCCAUAAAUUCAUAUUCCCAUCAUUCAAUUGACAAAUGGCCAUAUGUGGCCAGUGACAUGGUUUCUCCACCAGCUCCCCCCUC---------------CACAAGUGGAGGCA .(((((....((((.......(((......)))....((((((....)))))))))).))))).................((((---------------((....)))))).. ( -26.00) >DroYak_CAF1 11567 113 - 1 UGCCAUAAAUUCAUAUUCCCAUCAUUCAAUUGACAAAUGGCCAUAUGUGGCCAGUGACAUGGUUUCUCCGCCAUCGCCGCCAUCGCCGCCAUCGUCGCCAAUAAGUGGAGGCA .(((...........................(((..(((((.....(((((..((((..((((......)))))))).)))))....))))).))).(((.....))).))). ( -29.70) >DroAna_CAF1 11382 84 - 1 UGCCAUAAAUUCAUAUUCCCAUCAUUCAAUUGACAAAUGGCCAUAUGUGGCCAGUGACAUGGUUUCU--------ACCCCCACC---------------CGCAAGGA------ .(((((....((((.......(((......)))....((((((....)))))))))).)))))....--------........(---------------(....)).------ ( -20.50) >consensus UGCCAUAAAUUCAUAUUCCCAUCAUUCAAUUGACAAAUGGCCAUAUGUGGCCAGUGACAUGGUUUCUCCGCCAUCACCCCCGCC_______________CAUAAGUGGAGGCA .(((((....((((.......(((......)))....((((((....)))))))))).))))).(((((((.................................))))))).. (-19.46 = -20.39 + 0.93)

| Location | 1,614,179 – 1,614,284 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.70 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -12.36 |
| Energy contribution | -12.92 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.48 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
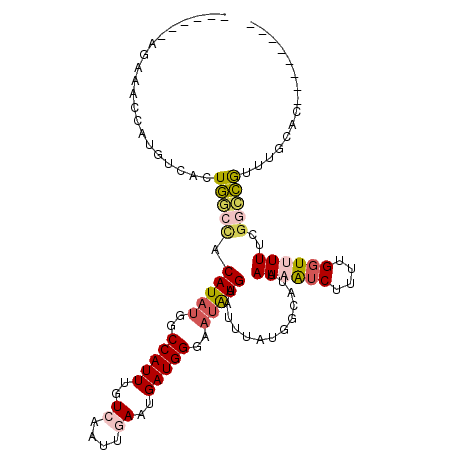
>X_DroMel_CAF1 1614179 105 + 22224390 UGGCGGAGAAACCAUGUCACUGGCCACAUAUGGCCAUUUGUCAAUUGAAUGAUGGGAAUAUGAAUUUAUGGCAUUAAAAUCUUUUGGUUUUUCGGCCGUUUGCAC-------- .(((.((((((((((((((.((((((....))))))((..(((......)))..))............)))))...........))))))))).)))........-------- ( -33.30) >DroVir_CAF1 15359 86 + 1 -----------------UACUCCCCACAUACGACCAUUUGUAAAUUGAAUGAUGGAUUUAUGAGCUUAUAGCAUUAA-GUCCUUUGGUUUUUGACAGAAUUAAA--------- -----------------...................(((((.....((((.(.(((((((...((.....))..)))-))))..).))))...)))))......--------- ( -13.00) >DroGri_CAF1 12318 87 + 1 -----------------CAUUCACUGCAUUUGACCAUUUGUAAAUUGAAUGAUGGAUAUGUGAGUUUAUAGCAUUAA-GUCGUUUGGUUUUUGGCUGGAUUGAAC-------- -----------------..((((....((((..(((.....((((..((((((....((((.........))))...-))))))..)))).)))..)))))))).-------- ( -16.50) >DroSim_CAF1 25817 104 + 1 UGGCGGAGAAACCAUGUCACUGGCCACAUAUGGCCAUUUGUCAAUUGAAUGAUGGGAAUAUGAAUUUAUGGCAUUAAAAUCUUUUG-UUUUUCGGCCGUUUGCAC-------- .(((.(((((((.((((((.((((((....))))))((..(((......)))..))............))))))...........)-)))))).)))........-------- ( -28.60) >DroEre_CAF1 10598 105 + 1 UGGUGGAGAAACCAUGUCACUGGCCACAUAUGGCCAUUUGUCAAUUGAAUGAUGGGAAUAUGAAUUUAUGGCAUUAAAAUCUUUUGGUUUUUCGGCCGUUUGCAC-------- .(((.((((((((((((((.((((((....))))))((..(((......)))..))............)))))...........))))))))).)))........-------- ( -32.00) >DroAna_CAF1 11399 107 + 1 ------AGAAACCAUGUCACUGGCCACAUAUGGCCAUUUGUCAAUUGAAUGAUGGGAAUAUGAAUUUAUGGCAUUAAAAUCUUUUGGUUUUUCGGCCGUUUGGCCAUUUGGAC ------.....(((......(((((((((((..(((((..((....))..)))))..))))).....(((((...((((((....))))))...))))).))))))..))).. ( -32.60) >consensus ______AGAAACCAUGUCACUGGCCACAUAUGGCCAUUUGUCAAUUGAAUGAUGGGAAUAUGAAUUUAUGGCAUUAAAAUCUUUUGGUUUUUCGGCCGUUUGCAC________ ....................(((((.(((((..(((((..((....))..)))))..))))).............((((((....))))))..)))))............... (-12.36 = -12.92 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:46 2006