
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,469,279 – 14,469,454 |
| Length | 175 |
| Max. P | 0.901187 |

| Location | 14,469,279 – 14,469,376 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -21.22 |
| Energy contribution | -21.00 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14469279 97 + 22224390 --------------------AAAAGGCUUUGAUUUCAAAUCAAUGUUGUUGGAUCUUGUAGAACGAAGAUUAGUAGGAGUUUCUUUUGACACGGGUCUUACGAUACUUGAUCCG---CUU --------------------...((((.(((((.....))))).......(((((..(((...((((((((..((((((...)))))).....)))))).)).)))..))))))---))) ( -22.00) >DroSim_CAF1 6433 116 + 1 NNNNNNNNNNNNNNNNNNNNAAAAGGCUUUGAUUUCUAAUCAAUGUU-UUGGAUCGUGUAGAACGAAGAUUAGUAGGAGUUUUUUUUGACACGGGUCUUACGAUACUUGAUCCG---CUU ......................(((((.(((((.....))))).)))-))((((((.(((...((((((((.((..(((....)))..))...)))))).)).))).)))))).---... ( -22.40) >DroYak_CAF1 2533 100 + 1 --------------------AAAAGGCUUUGAUUUCUAAUCAAUGCUGUUGGAUCUUGUAGAACGAAGAUUAGUAGGAGUUUCUUUUGACACGGGUCUUACGAUACUUGAUCCGAUACUU --------------------....(((.(((((.....))))).)))((((((((..(((...((((((((..((((((...)))))).....)))))).)).)))..)))))))).... ( -26.60) >consensus ____________________AAAAGGCUUUGAUUUCUAAUCAAUGUUGUUGGAUCUUGUAGAACGAAGAUUAGUAGGAGUUUCUUUUGACACGGGUCUUACGAUACUUGAUCCG___CUU ........................(((.(((((.....))))).)))...(((((..(((...(((((((..((....(((......)))))..))))).)).)))..)))))....... (-21.22 = -21.00 + -0.22)


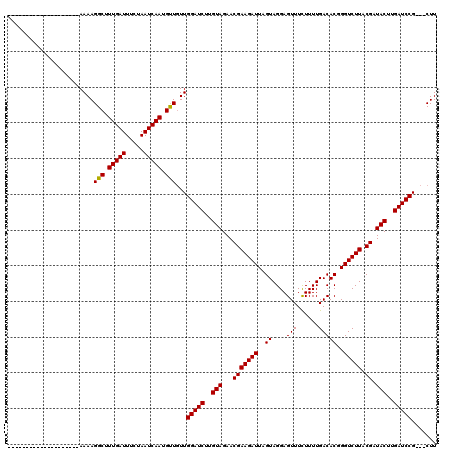
| Location | 14,469,339 – 14,469,454 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.16 |
| Mean single sequence MFE | -35.97 |
| Consensus MFE | -23.40 |
| Energy contribution | -23.02 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14469339 115 - 22224390 CAUUGACAACUUCAUUGGUGAAGUGAGGGGAAA-UGUCUUUCCGA-AAAGAACACAACUUUUGGGAAGGAGGCUCCAGAGAAG---CGGAUCAAGUAUCGUAAGACCCGUGUCAAAAGAA ..((((((((((((....))))))..(((....-..(((((((.(-((((.......))))).)))))))((.(((.......---.))))).....((....))))).))))))..... ( -33.80) >DroSec_CAF1 12 113 - 1 CAUUGACAACUUCAUCGGGGA--UUGGGGGAAA-UGUCUUCCCGA-AAAGAACACAACUUUUUGGGAGGAGGGUCUGGAGAAG---CGGAUCAAGUAUCGUAAGACCCGUGUCAAAAGAA ..((((((.(((((.......--.)))))....-..(((((((((-((((.......)))))))))))))((((((...((.(---(.......)).))...)))))).))))))..... ( -40.80) >DroSim_CAF1 6512 113 - 1 CAUUGACAACUUCAUCGGGGA--UUGGGGGAAA-UGUCUUCUCCA-AAAGAACACAACUUUUUGGGAGGAGGGUCUAGAGAAG---CGGAUCAAGUAUCGUAAGACCCGUGUCAAAAAAA ..((((((.(((((.......--.)))))....-..(((((.(((-((((........))))))))))))((((((...((.(---(.......)).))...)))))).))))))..... ( -33.50) >DroYak_CAF1 2593 119 - 1 CAUUGACAACUUCAUCAGGGGA-GUGGAGGGAGUUCGUUUCCCGAAAAACAAGACAACUUUUUGGGAGGAGGGUCCCGAGAAGUAUCGGAUCAAGUAUCGUAAGACCCGUGUCAAAAGAA ..((((((((((.(((.(((((-(((((.....)))))))))).........((..(((((((((((......))))))))))).)).))).)))).((....))....))))))..... ( -35.80) >consensus CAUUGACAACUUCAUCGGGGA__UUGGGGGAAA_UGUCUUCCCGA_AAAGAACACAACUUUUUGGGAGGAGGGUCCAGAGAAG___CGGAUCAAGUAUCGUAAGACCCGUGUCAAAAGAA ..((((((((((.(((.(..........(((...(.(((((((((.((((.......))))))))))))).).)))..........).))).)))).((....))....))))))..... (-23.40 = -23.02 + -0.37)

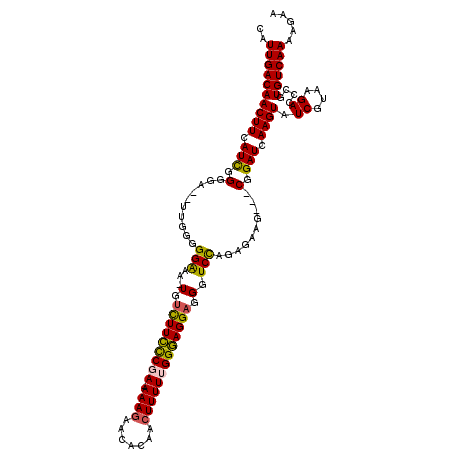

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:12 2006