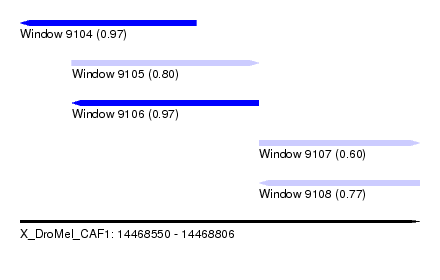
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,468,550 – 14,468,806 |
| Length | 256 |
| Max. P | 0.971993 |

| Location | 14,468,550 – 14,468,663 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.77 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -35.47 |
| Energy contribution | -35.37 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14468550 113 - 22224390 ACGCCAAAGCGCAAUCAAAAAGCGUUUUUAAUAAUGCGCUAAGUGAUUGUUAAGCGCUGGUGGAGAUGGAGGUCCCAACUCGAUGGCUUUCCCAAUUCCACAGCUCACA-------GAUU ........(((((((((...(((((.((....)).)))))...)))))))...))((((.(((((.(((..(..(((......)))..)..))).))))))))).....-------.... ( -35.30) >DroSim_CAF1 5237 113 - 1 ACGCCAAAGCGCAAUCAAAAAGCGUUUUUAAUAAUGCGCUAAGUGAUUGUUAAGCGCUGGUGGAGAUGGAGGGCCCAACUCGAUGGCUUUCCCAAUCCCACAGCUCACA-------GAUU ........(((((((((...(((((.((....)).)))))...)))))))...))((((..(((...((((((((.........))))))))...)))..)))).....-------.... ( -38.00) >DroYak_CAF1 1753 120 - 1 ACGCCAAAGCGCAAUCAAAAAGCGUUUUUAAUAAUGCGCUAAGUGAUUGUUAAGCGCUGGCGGAGAUGGAGAGCCCAACUCGAUGGCUUUCCCAAUCCCACAGCUCACAGCUCACGGAUU .(((((..(((((((((...(((((.((....)).)))))...)))))))...))..))))).....((((((((.........)))))))).(((((...(((.....)))...))))) ( -43.40) >consensus ACGCCAAAGCGCAAUCAAAAAGCGUUUUUAAUAAUGCGCUAAGUGAUUGUUAAGCGCUGGUGGAGAUGGAGGGCCCAACUCGAUGGCUUUCCCAAUCCCACAGCUCACA_______GAUU ........(((((((((...(((((.((....)).)))))...)))))))...))((((..(((...((((((((.........))))))))...)))..))))................ (-35.47 = -35.37 + -0.11)

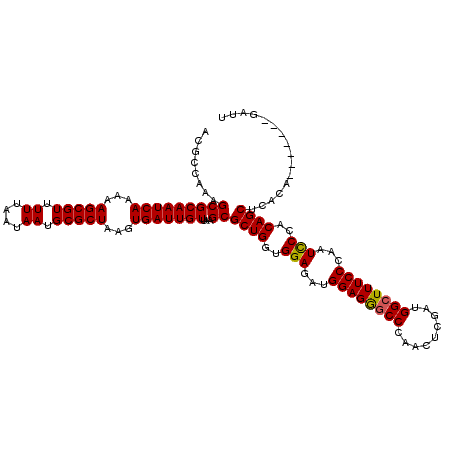
| Location | 14,468,583 – 14,468,703 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -38.63 |
| Consensus MFE | -35.69 |
| Energy contribution | -35.80 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14468583 120 + 22224390 AGUUGGGACCUCCAUCUCCACCAGCGCUUAACAAUCACUUAGCGCAUUAUUAAAAACGCUUUUUGAUUGCGCUUUGGCGUGUUGGCCCAUUGAAAUCGAUAGGCGUUGCCCGGUGGAUCU .....((....))...((((((.(((((............))))).........(((((((.((((((...(..(((.((....)))))..).)))))).)))))))....))))))... ( -38.20) >DroSim_CAF1 5270 120 + 1 AGUUGGGCCCUCCAUCUCCACCAGCGCUUAACAAUCACUUAGCGCAUUAUUAAAAACGCUUUUUGAUUGCGCUUUGGCGUGUUGGCCCAUUGAAAUCGAUAGGCGUUGCCCGGUGGAUCU .(.(((.....))).)((((((.(((((............))))).........(((((((.((((((...(..(((.((....)))))..).)))))).)))))))....))))))... ( -38.10) >DroYak_CAF1 1793 120 + 1 AGUUGGGCUCUCCAUCUCCGCCAGCGCUUAACAAUCACUUAGCGCAUUAUUAAAAACGCUUUUUGAUUGCGCUUUGGCGUGUUGGCCCAUUGAAAUCGAUAGGCGUAGCCCAGUGGAUCU ..((((((((.((......((((((((....((((((...((((............))))...)))))).((....))))))))))..((((....)))).)).).)))))))....... ( -39.60) >consensus AGUUGGGCCCUCCAUCUCCACCAGCGCUUAACAAUCACUUAGCGCAUUAUUAAAAACGCUUUUUGAUUGCGCUUUGGCGUGUUGGCCCAUUGAAAUCGAUAGGCGUUGCCCGGUGGAUCU .(.(((.....))).)((((((.(.((.(((((....(..((((((..(((((((.....)))))))))))))..)...)))))(((.((((....)))).)))...))).))))))... (-35.69 = -35.80 + 0.11)

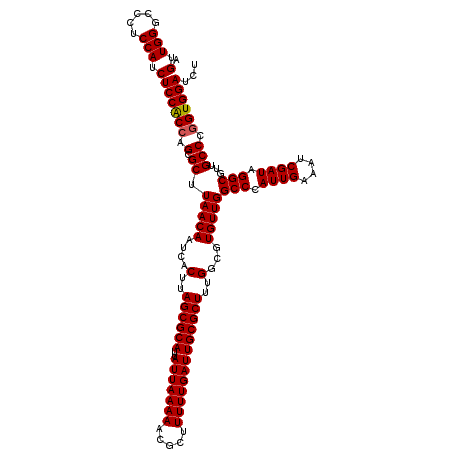
| Location | 14,468,583 – 14,468,703 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -41.67 |
| Consensus MFE | -38.45 |
| Energy contribution | -38.57 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14468583 120 - 22224390 AGAUCCACCGGGCAACGCCUAUCGAUUUCAAUGGGCCAACACGCCAAAGCGCAAUCAAAAAGCGUUUUUAAUAAUGCGCUAAGUGAUUGUUAAGCGCUGGUGGAGAUGGAGGUCCCAACU ...((((((((((...((((((.(....).))))))......((....))(((((((...(((((.((....)).)))))...)))))))...)).))))))))..(((.....)))... ( -40.80) >DroSim_CAF1 5270 120 - 1 AGAUCCACCGGGCAACGCCUAUCGAUUUCAAUGGGCCAACACGCCAAAGCGCAAUCAAAAAGCGUUUUUAAUAAUGCGCUAAGUGAUUGUUAAGCGCUGGUGGAGAUGGAGGGCCCAACU ...((((((((((...((((((.(....).))))))......((....))(((((((...(((((.((....)).)))))...)))))))...)).))))))))..(((.....)))... ( -40.80) >DroYak_CAF1 1793 120 - 1 AGAUCCACUGGGCUACGCCUAUCGAUUUCAAUGGGCCAACACGCCAAAGCGCAAUCAAAAAGCGUUUUUAAUAAUGCGCUAAGUGAUUGUUAAGCGCUGGCGGAGAUGGAGAGCCCAACU ........((((((..((((((.(....).)))))).....(((((..(((((((((...(((((.((....)).)))))...)))))))...))..))))).........))))))... ( -43.40) >consensus AGAUCCACCGGGCAACGCCUAUCGAUUUCAAUGGGCCAACACGCCAAAGCGCAAUCAAAAAGCGUUUUUAAUAAUGCGCUAAGUGAUUGUUAAGCGCUGGUGGAGAUGGAGGGCCCAACU .........((((...((((((.(....).)))))).....(((((..(((((((((...(((((.((....)).)))))...)))))))...))..)))))..........)))).... (-38.45 = -38.57 + 0.11)



| Location | 14,468,703 – 14,468,806 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -15.42 |
| Energy contribution | -15.76 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.595779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14468703 103 + 22224390 UGGCCAGCGGCUUAGCAUUAGAUACCCUAACUGCGAAGAUCUUGACAGGGUAUCGUAUUAUUUUCAACCAAUACAAAUACAU------------UGUAAGUUAAGAUAAUACUU-----A .((((...))))........((((((((.....(((.....)))..))))))))((((((((((.(((...(((((.....)------------)))).)))))))))))))..-----. ( -25.40) >DroSim_CAF1 5390 114 + 1 UGGCCAGCGGCUUAGCAUUAGAUACCCUAACUGCGGAGAUCUUGACAGGGUAUCGUAUUAUUUU-AAGUAAUAAAAAUACAUGUUACUUCAAGGUCUAAGUUAAAAUAAUACCA-----A .((((...))))........((((((((..(...((....)).)..))))))))((((((((((-((((((((........))))))).............)))))))))))..-----. ( -24.71) >DroYak_CAF1 1913 116 + 1 UGGCCAGCGGCUGAGCGUUAGAUACCCUAGCUCCACAGAUCUUGACAGGGUAUCAUAGUAUUUC-AAUCUAUAAAAAUAUAUCGUACUUAAAC---GAAAUUAAAAUAGUUUCAUCGGAA .((((...))))(((((.((((((((((..(............)..))))))))((((......-...)))).......)).)))........---(((((((...))))))).)).... ( -20.90) >consensus UGGCCAGCGGCUUAGCAUUAGAUACCCUAACUGCGAAGAUCUUGACAGGGUAUCGUAUUAUUUU_AACCAAUAAAAAUACAU__UACUU_AA__U_UAAGUUAAAAUAAUACCA_____A .((((...))))........((((((((..(............)..))))))))((((((((((......................................))))))))))........ (-15.42 = -15.76 + 0.34)


| Location | 14,468,703 – 14,468,806 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -17.01 |
| Energy contribution | -17.24 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14468703 103 - 22224390 U-----AAGUAUUAUCUUAACUUACA------------AUGUAUUUGUAUUGGUUGAAAAUAAUACGAUACCCUGUCAAGAUCUUCGCAGUUAGGGUAUCUAAUGCUAAGCCGCUGGCCA .-----..(((((((.((((((((((------------(.....)))))..))))))..)))))))(((((((((....(.....).....)))))))))....((((......)))).. ( -27.00) >DroSim_CAF1 5390 114 - 1 U-----UGGUAUUAUUUUAACUUAGACCUUGAAGUAACAUGUAUUUUUAUUACUU-AAAAUAAUACGAUACCCUGUCAAGAUCUCCGCAGUUAGGGUAUCUAAUGCUAAGCCGCUGGCCA .-----..(((((((((((((((........)))).....(((.......))).)-))))))))))(((((((((....(.....).....)))))))))....((((......)))).. ( -24.80) >DroYak_CAF1 1913 116 - 1 UUCCGAUGAAACUAUUUUAAUUUC---GUUUAAGUACGAUAUAUUUUUAUAGAUU-GAAAUACUAUGAUACCCUGUCAAGAUCUGUGGAGCUAGGGUAUCUAACGCUCAGCCGCUGGCCA ......(((...((((((((((((---((......))))((((....))))))))-))))))....(((((((((......((....))..)))))))))......)))(((...))).. ( -22.50) >consensus U_____UGGUAUUAUUUUAACUUA_A__UU_AAGUA__AUGUAUUUUUAUUGAUU_AAAAUAAUACGAUACCCUGUCAAGAUCUCCGCAGUUAGGGUAUCUAAUGCUAAGCCGCUGGCCA ........((((((((((((((.............................)))).))))))))))(((((((((................))))))))).........(((...))).. (-17.01 = -17.24 + 0.23)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:10 2006