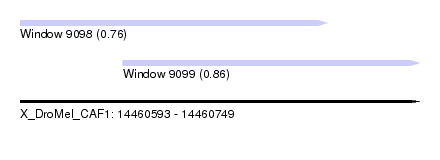
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,460,593 – 14,460,749 |
| Length | 156 |
| Max. P | 0.864296 |

| Location | 14,460,593 – 14,460,713 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.94 |
| Mean single sequence MFE | -22.63 |
| Consensus MFE | -14.69 |
| Energy contribution | -15.47 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14460593 120 + 22224390 UUUCCUAAUUAAGUCGCCCAGCCAAAAAAAAAAAAAAACACACACACGCACUCAUUCGCCUGCAAAGUGGGCCCGAAAUAAAAUCAUUUAACAAAAUAAGUUAAGUGAACGGGGACUCUA ...........((((.(((......................................(((..(...)..)))...........(((((((((.......)))))))))..)))))))... ( -24.60) >DroEre_CAF1 5491 107 + 1 UUUCCUAAUUAAGUGGCCCAGCCAAAAAA-----CACGC--------ACACUCAUUCGCCUGGAAAGUGGCCCCAAAAUAAAAUCAUUUAACAAAAUAAGUUAAGUGAACGGGGAUGCUA ...........(((..(((.((((.....-----((.((--------..........)).)).....))))............(((((((((.......)))))))))...)))..))). ( -24.00) >DroYak_CAF1 6461 115 + 1 UUUCCUAAUUAAGUCGCCCAGUCAAAAAA-----CACACACACACACACACUUAUUUGCCUGAAAAGUGGGCCCAAAAUAAAAUCAUUUAACAAAAUAAGUUAAGUGCACGGGGAUUAUA ...........((((.(((..........-----.................((((((((((.......))))...))))))...((((((((.......))))))))...)))))))... ( -19.30) >consensus UUUCCUAAUUAAGUCGCCCAGCCAAAAAA_____CACACACACACACACACUCAUUCGCCUGAAAAGUGGGCCCAAAAUAAAAUCAUUUAACAAAAUAAGUUAAGUGAACGGGGAUUCUA ............(((.(((......................................((((.......))))...........(((((((((.......)))))))))..)))))).... (-14.69 = -15.47 + 0.78)

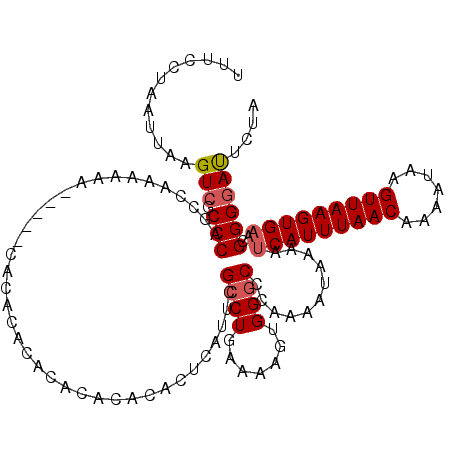
| Location | 14,460,633 – 14,460,749 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.11 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -22.60 |
| Energy contribution | -22.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14460633 116 + 22224390 CACACACGCACUCAUUCGCCUGCAAAGUGGGCCCGAAAUAAAAUCAUUUAACAAAAUAAGUUAAGUGAACGGGGACUCUAAAUGGCAGGCACUAUUCACUAAAGAUUGUGAUCUAA---- .................((((((....((((((((........(((((((((.......))))))))).))))...))))....)))))).....((((........)))).....---- ( -30.10) >DroEre_CAF1 5525 113 + 1 -------ACACUCAUUCGCCUGGAAAGUGGCCCCAAAAUAAAAUCAUUUAACAAAAUAAGUUAAGUGAACGGGGAUGCUAAAUGGCGGGCACUAUUCACUAAAGAGUGUGAUGUAUAGUG -------.......((((((.....(((..((((.........(((((((((.......)))))))))..))))..)))....))))))((((((((((........))))...)))))) ( -31.00) >DroYak_CAF1 6496 116 + 1 CACACACACACUUAUUUGCCUGAAAAGUGGGCCCAAAAUAAAAUCAUUUAACAAAAUAAGUUAAGUGCACGGGGAUUAUAAAUGGCAGGCACUAUUCACUAAAGAUUGUGAUGCAU---- ................((((((....((((.(((..........((((((((.......))))))))....))).))))......))))))....((((........)))).....---- ( -22.14) >consensus CACACACACACUCAUUCGCCUGAAAAGUGGGCCCAAAAUAAAAUCAUUUAACAAAAUAAGUUAAGUGAACGGGGAUUCUAAAUGGCAGGCACUAUUCACUAAAGAUUGUGAUGUAU____ .................(((((.....(((((((.........(((((((((.......)))))))))...)))..)))).....))))).....((((........))))......... (-22.60 = -22.60 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:02 2006