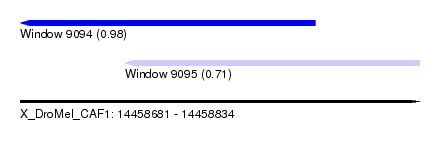
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,458,681 – 14,458,834 |
| Length | 153 |
| Max. P | 0.980432 |

| Location | 14,458,681 – 14,458,794 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.32 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -19.52 |
| Energy contribution | -19.87 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14458681 113 - 22224390 UUCG-----AUUUUUUUGUACGCCGAUGAACAAUGAUUUCGCAUUGUUUGUUUGUGUGCAAAAACUCAAAUUAUUAAUUACCAAGCCAGAAUGUUUUUGUUUUUCGUGAUUUUCUUUG ...(-----(..(((((((((((.((..(((((((......)))))))..)).))))))))))).))(((((((.((..((.((((......))))..))..)).)))))))...... ( -27.40) >DroEre_CAF1 3601 104 - 1 CUCACU-----UUUUUG-CUCGCCGAUGAACAAUGAUUUCGCAUUGUUCG---GCGGGCGAAAACUGAAAUUAUUAAUUACCAAGCAAGAAUGUUUUUGUUUUUCGUGAUUUU----- .(((..-----((((((-((((((((...((((((......)))))))))---))))))))))).))).......((((((.((((((((....))))))))...))))))..----- ( -34.10) >DroYak_CAF1 4574 112 - 1 CUCACUCACAUUUUUUGUUUCGCCGAUGAACAAUGAUUUCGCCUUGUUCG---GUGUGCAAAAACUGAAAUUAUUAAUUACCCGGCAAGAAUGUUUUUGUUUUUCGUGAUUUUCU--- .((((...((.(((((((..((((((...((((.(......).)))))))---))).))))))).))................(((((((....)))))))....))))......--- ( -22.30) >consensus CUCACU___AUUUUUUG_UUCGCCGAUGAACAAUGAUUUCGCAUUGUUCG___GUGUGCAAAAACUGAAAUUAUUAAUUACCAAGCAAGAAUGUUUUUGUUUUUCGUGAUUUUCU___ ...........((((((.(((((...(((((((((......)))))))))...)))))))))))...........((((((.((((((((....))))))))...))))))....... (-19.52 = -19.87 + 0.35)


| Location | 14,458,721 – 14,458,834 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 85.92 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -21.44 |
| Energy contribution | -20.90 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14458721 113 - 22224390 CGCGAUGAGAUGACAUAAUUGCUUGGCUUCUUGUGAAUCAUUCG-----AUUUUUUUGUACGCCGAUGAACAAUGAUUUCGCAUUGUUUGUUUGUGUGCAAAAACUCAAAUUAUUAAU .....((((.......(((((..(((.(((....))))))..))-----)))(((((((((((.((..(((((((......)))))))..)).))))))))))))))).......... ( -32.20) >DroEre_CAF1 3636 108 - 1 CGCGAUGAGAUGACAUAAUUGC-CGGCUUCUUGUGAAUCACUCACU-----UUUUUG-CUCGCCGAUGAACAAUGAUUUCGCAUUGUUCG---GCGGGCGAAAACUGAAAUUAUUAAU (((((.(((.((.((....)).-)).))).)))))......(((..-----((((((-((((((((...((((((......)))))))))---))))))))))).))).......... ( -34.50) >DroYak_CAF1 4611 115 - 1 CGCGAUGAGAUGACAUAAUUGCUCGGCUUCUUGUGAAUCACUCACUCACAUUUUUUGUUUCGCCGAUGAACAAUGAUUUCGCCUUGUUCG---GUGUGCAAAAACUGAAAUUAUUAAU ...((((((.((((((((..((...))...)))))..))))))).)).((.(((((((..((((((...((((.(......).)))))))---))).))))))).))........... ( -24.70) >consensus CGCGAUGAGAUGACAUAAUUGCUCGGCUUCUUGUGAAUCACUCACU___AUUUUUUG_UUCGCCGAUGAACAAUGAUUUCGCAUUGUUCG___GUGUGCAAAAACUGAAAUUAUUAAU .....((((.((((((((..((...))...)))))..))))))).......((((((.(((((...(((((((((......)))))))))...))))))))))).............. (-21.44 = -20.90 + -0.54)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:58 2006