
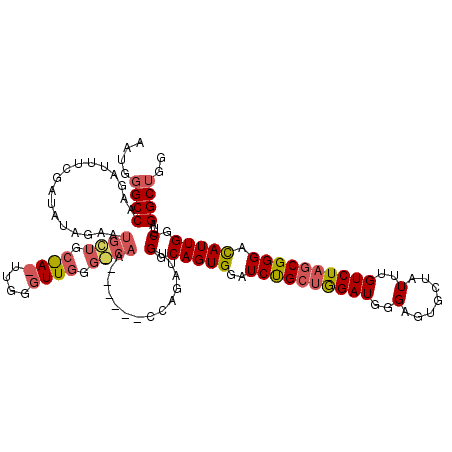
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,454,987 – 14,455,107 |
| Length | 120 |
| Max. P | 0.940604 |

| Location | 14,454,987 – 14,455,107 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -37.27 |
| Consensus MFE | -25.17 |
| Energy contribution | -27.43 |
| Covariance contribution | 2.26 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14454987 120 + 22224390 AAUGGGCCAAGAUUUCGAUAUAGAAUGCUGCAAAUUGGGUUGGGGCAAUGGAACCCAGAUUGGCAGUGGAUCUGCUAGAUGGGAGUGCUAUUUGUCUAGCGGGACAUUGGCUUAGGCUGG ..((((((((..(..((...((((.((((.(((......))).))))((((..((((..(((((((.....))))))).))))....))))...)))).))..)..))))))))...... ( -41.70) >DroSim_CAF1 762 113 + 1 AAUGGGCCAAGAUUUCGAUCUAGAAUGGUGCGACUUGGGUUGGGCCAA-------CAGAUUGGCAGUGGAUCUGCUGGAUGGGAGUGCUAUUUGUCUAGCGGGAUAUUGGCUUAGGCUGG ....((((...(((((.((((((..((((.((((....)))).)))).-------((((((.......)))))))))))).)))))((((..(((((....))))).))))...)))).. ( -40.30) >DroEre_CAF1 350 95 + 1 AACGUGCCAAGAUU-GGAUACAGAAUGCUGC-----------------------CCAGAUUGGCAGUGGAUCCG-CGGAUGGGAGUGCUAUUUGUCUAGCGGGACAUUGGCUUAGGCUGG ..((.(((......-((((......((((((-----------------------(......))))))).))))(-(.((((....(((((......)))))...)))).))...))))). ( -29.80) >consensus AAUGGGCCAAGAUUUCGAUAUAGAAUGCUGC_A_UUGGGUUGGG_CAA______CCAGAUUGGCAGUGGAUCUGCUGGAUGGGAGUGCUAUUUGUCUAGCGGGACAUUGGCUUAGGCUGG ....((((.................((((.((((....)))).))))..............(.(((((..((((((((((..(.......)..)))))))))).))))).)...)))).. (-25.17 = -27.43 + 2.26)


| Location | 14,454,987 – 14,455,107 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -13.59 |
| Energy contribution | -13.37 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14454987 120 - 22224390 CCAGCCUAAGCCAAUGUCCCGCUAGACAAAUAGCACUCCCAUCUAGCAGAUCCACUGCCAAUCUGGGUUCCAUUGCCCCAACCCAAUUUGCAGCAUUCUAUAUCGAAAUCUUGGCCCAUU .........(((((.((..(((((((...............))))))(((....((((.(((.((((((..........))))))))).))))...))).....)..)).)))))..... ( -26.16) >DroSim_CAF1 762 113 - 1 CCAGCCUAAGCCAAUAUCCCGCUAGACAAAUAGCACUCCCAUCCAGCAGAUCCACUGCCAAUCUG-------UUGGCCCAACCCAAGUCGCACCAUUCUAGAUCGAAAUCUUGGCCCAUU .........(((((.((..(((((((......(((((.....(((((((((.........)))))-------)))).........))).)).....)))))..))..)).)))))..... ( -22.84) >DroEre_CAF1 350 95 - 1 CCAGCCUAAGCCAAUGUCCCGCUAGACAAAUAGCACUCCCAUCCG-CGGAUCCACUGCCAAUCUGG-----------------------GCAGCAUUCUGUAUCC-AAUCUUGGCACGUU .........(((((......((((......))))..........(-((((....(((((......)-----------------------))))...)))))....-....)))))..... ( -24.80) >consensus CCAGCCUAAGCCAAUGUCCCGCUAGACAAAUAGCACUCCCAUCCAGCAGAUCCACUGCCAAUCUGG______UUG_CCCAACCCAA_U_GCAGCAUUCUAUAUCGAAAUCUUGGCCCAUU .........(((((......((((......))))............(((((.........))))).............................................)))))..... (-13.59 = -13.37 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:57 2006