
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,452,927 – 14,453,017 |
| Length | 90 |
| Max. P | 0.908276 |

| Location | 14,452,927 – 14,453,017 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 90.84 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.56 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
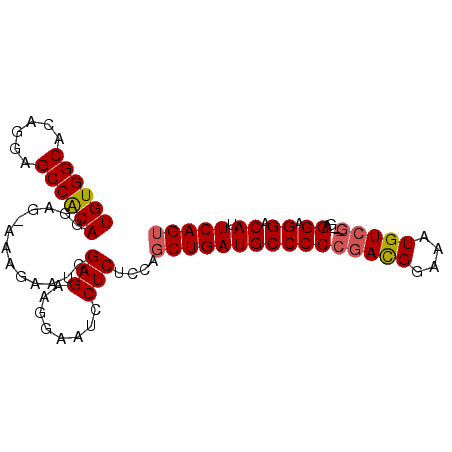
>X_DroMel_CAF1 14452927 90 + 22224390 ACUGAAUGUCCUCCUC---CGACAUUUCCGUCGGGGGCAUCAGCUGUUGAGGAUUCCUUCUCGAUUCUUUCCUCCUGUGGCUCCUGUGCCACA .((((.(((((....(---((((......))))))))))))))..((((((((....))))))))...........(((((......))))). ( -32.20) >DroSec_CAF1 13970 89 + 1 ACUGAAUGUCCUCCUG---CGACACUUCCGUCGGGGGCAUCAGCUGGAGAGGAUUCCUUCUCGAUUCUUU-CUCCUGUGGCUCCUGUGCCACA .((((.(((((.((.(---((.......))).)))))))))))..((((((((.((......)).)).))-)))).(((((......))))). ( -35.10) >DroEre_CAF1 14933 89 + 1 ACUGAAUGUCCUCCUC---CGACAUUUCCGUCGGGGGCAUCAGCUGAAGAGGAUUCCUUCUCGAUUCCUU-CUCCUGCGGCUCCUGUGCCACA ..(((.(((((....(---((((......)))))))))))))((.((..((((.((......)).)))).-.))..))(((......)))... ( -30.80) >DroYak_CAF1 12936 92 + 1 UCUGAAUGUCCUCCUCCUCUGACAUUUCCAUUGGGGGCAUCAGCUGGCGAGGAUUCCUUCUCGAUUCUUU-CUCAUGCGGCUCCUGUGCCACA ...(((.((((((..((.((((..(..((...))..)..))))..)).))))))...)))..........-....((.(((......))).)) ( -23.30) >consensus ACUGAAUGUCCUCCUC___CGACAUUUCCGUCGGGGGCAUCAGCUGGAGAGGAUUCCUUCUCGAUUCUUU_CUCCUGCGGCUCCUGUGCCACA .((((.(((((((.......(((......)))))))))))))).....(((((....))))).............((((((......)))))) (-23.00 = -23.56 + 0.56)



| Location | 14,452,927 – 14,453,017 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 90.84 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -21.99 |
| Energy contribution | -22.30 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14452927 90 - 22224390 UGUGGCACAGGAGCCACAGGAGGAAAGAAUCGAGAAGGAAUCCUCAACAGCUGAUGCCCCCGACGGAAAUGUCG---GAGGAGGACAUUCAGU ((((((......)))))).(((((................)))))....(((((..((.((((((....)))))---).)).......))))) ( -29.79) >DroSec_CAF1 13970 89 - 1 UGUGGCACAGGAGCCACAGGAG-AAAGAAUCGAGAAGGAAUCCUCUCCAGCUGAUGCCCCCGACGGAAGUGUCG---CAGGAGGACAUUCAGU ((((((......))))))((((-(..((.((......)).)).))))).((((((((((((((((....)))))---..)).)).)).))))) ( -32.10) >DroEre_CAF1 14933 89 - 1 UGUGGCACAGGAGCCGCAGGAG-AAGGAAUCGAGAAGGAAUCCUCUUCAGCUGAUGCCCCCGACGGAAAUGUCG---GAGGAGGACAUUCAGU ((((((......))))))((((-(.(((.((......)).)))))))).(((((..((.((((((....)))))---).)).......))))) ( -34.30) >DroYak_CAF1 12936 92 - 1 UGUGGCACAGGAGCCGCAUGAG-AAAGAAUCGAGAAGGAAUCCUCGCCAGCUGAUGCCCCCAAUGGAAAUGUCAGAGGAGGAGGACAUUCAGA ((((((......))))))....-..........(((....(((((.((..(((((.((......))....))))).))..)))))..)))... ( -25.20) >consensus UGUGGCACAGGAGCCACAGGAG_AAAGAAUCGAGAAGGAAUCCUCUCCAGCUGAUGCCCCCGACGGAAAUGUCG___GAGGAGGACAUUCAGU ((((((......)))))).............(((........)))....((((((((((((((((....))))).....)).)).)).))))) (-21.99 = -22.30 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:54 2006