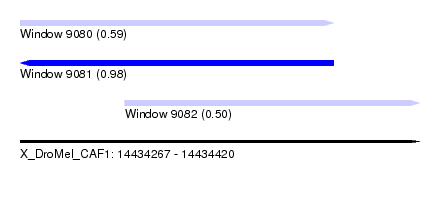
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,434,267 – 14,434,420 |
| Length | 153 |
| Max. P | 0.984082 |

| Location | 14,434,267 – 14,434,387 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -25.34 |
| Energy contribution | -26.22 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14434267 120 + 22224390 GUUCUAAAUUAUUAGCUAAAUGCAUCCGAUCAUCAGUCGCAAACUAUUUUCGCCUGUGGACAAUAAGCGAGCAAGUGCCGCAAUUGCUGACCAGGCGGACUAAUAGAUGCGAAUUUAAAU ..............((.....)).............(((((..(((((((((((((.((.((((..(((.((....))))).))))))...))))))))..))))).)))))........ ( -31.10) >DroSec_CAF1 13632 120 + 1 GUUCUAAAUUAUUAGCUAAAUGCAUCCGAUCAUCAGUUGCAAACUAUUUUCGCCUGUGGACAAUAAGCGAGCAAGUGCCGCAAUUGCUGACCAGGCGAACUAAUAGAUGCGAUUUUAAAU ..............((.....))...........(((((((..(((((((((((((.((.((((..(((.((....))))).))))))...))))))))..))))).)))))))...... ( -30.90) >DroSim_CAF1 24881 120 + 1 GUUCUAAAUUAUUAGCUAAAUGCAUCCGAUCAUCAGUGGCAAACUAUUUUCGCCUGUGGACAAUAAGCGAGCAAGUGCCGCAAUUGCUGACCAGGCGAACUAAUAGAUGCGAAUUUAAAU ....((((((....((.....))...............(((..(((((((((((((.((.((((..(((.((....))))).))))))...))))))))..))))).))).))))))... ( -29.50) >DroEre_CAF1 13386 116 + 1 GUUCUAAAUUAUUAGCUAAAUGCAUCCGAUCAUCAGUCGCAAGCUAUUUUCGCCUGUGGACAAUGAGCGAGCCAGUGCCGCAAUUGCUA----GUGCCUAUAGUAGAUGCGAAUUUAAAU ....((((((...........(((((((.((((...(((((.((.......)).)))))...)))).)).)...))))((((.((((((----.......)))))).))))))))))... ( -25.20) >DroYak_CAF1 13637 120 + 1 GUUCUAAAUUAUUAGCUAAAUGCAUCCGAUCAUCAGUCGCAAACUAUUUUCGCUUGCGGACAAUAAGCGAGCGAGUGCCGCAACUGCUGAGCAGGCGGAAUAAUAGAUGCGAAUUUAAAU ..............((.....)).............(((((..((((((((((((((.........)))))))))..((((..((((...))))))))...))))).)))))........ ( -32.50) >consensus GUUCUAAAUUAUUAGCUAAAUGCAUCCGAUCAUCAGUCGCAAACUAUUUUCGCCUGUGGACAAUAAGCGAGCAAGUGCCGCAAUUGCUGACCAGGCGAACUAAUAGAUGCGAAUUUAAAU ..............((.....)).............(((((..((((((((((((((((.((((..(((.((....))))).)))))))..))))))))..))))).)))))........ (-25.34 = -26.22 + 0.88)



| Location | 14,434,267 – 14,434,387 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -27.12 |
| Energy contribution | -28.28 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14434267 120 - 22224390 AUUUAAAUUCGCAUCUAUUAGUCCGCCUGGUCAGCAAUUGCGGCACUUGCUCGCUUAUUGUCCACAGGCGAAAAUAGUUUGCGACUGAUGAUCGGAUGCAUUUAGCUAAUAAUUUAGAAC ..........(((((((((((((((((((....(((((.(((((....)).)))..)))))...))))))......(....))))))))....))))))......((((....))))... ( -36.60) >DroSec_CAF1 13632 120 - 1 AUUUAAAAUCGCAUCUAUUAGUUCGCCUGGUCAGCAAUUGCGGCACUUGCUCGCUUAUUGUCCACAGGCGAAAAUAGUUUGCAACUGAUGAUCGGAUGCAUUUAGCUAAUAAUUUAGAAC .............((((....((((((((....(((((.(((((....)).)))..)))))...))))))))..(((((((((.(((.....))).))))...)))))......)))).. ( -35.80) >DroSim_CAF1 24881 120 - 1 AUUUAAAUUCGCAUCUAUUAGUUCGCCUGGUCAGCAAUUGCGGCACUUGCUCGCUUAUUGUCCACAGGCGAAAAUAGUUUGCCACUGAUGAUCGGAUGCAUUUAGCUAAUAAUUUAGAAC ..........((((((((((.((((((((....(((((.(((((....)).)))..)))))...))))))))..((((.....)))).)))).))))))......((((....))))... ( -35.50) >DroEre_CAF1 13386 116 - 1 AUUUAAAUUCGCAUCUACUAUAGGCAC----UAGCAAUUGCGGCACUGGCUCGCUCAUUGUCCACAGGCGAAAAUAGCUUGCGACUGAUGAUCGGAUGCAUUUAGCUAAUAAUUUAGAAC ..........((((((.......((..----..))....(((((....)).)))((((((((..(((((.......))))).))).)))))..))))))......((((....))))... ( -28.00) >DroYak_CAF1 13637 120 - 1 AUUUAAAUUCGCAUCUAUUAUUCCGCCUGCUCAGCAGUUGCGGCACUCGCUCGCUUAUUGUCCGCAAGCGAAAAUAGUUUGCGACUGAUGAUCGGAUGCAUUUAGCUAAUAAUUUAGAAC ..........(((((((((((.((((((((...))))..))))((.((((..(((.(((.(((....).)).))))))..)))).))))))).))))))......((((....))))... ( -32.60) >consensus AUUUAAAUUCGCAUCUAUUAGUCCGCCUGGUCAGCAAUUGCGGCACUUGCUCGCUUAUUGUCCACAGGCGAAAAUAGUUUGCGACUGAUGAUCGGAUGCAUUUAGCUAAUAAUUUAGAAC .((((((((..............((((((....(((((.(((((....)).)))..)))))...))))))....(((((((((.(((.....))).))))...)))))..)))))))).. (-27.12 = -28.28 + 1.16)


| Location | 14,434,307 – 14,434,420 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.36 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -22.52 |
| Energy contribution | -24.00 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14434307 113 + 22224390 AAACUAUUUUCGCCUGUGGACAAUAAGCGAGCAAGUGCCGCAAUUGCUGACCAGGCGGACUAAUAGAUGCGAAUUUAAAUGCGCUCUCAAGAUUCUAUAUCUGCAGCAUUC-------GU ...(((((((((((((.((.((((..(((.((....))))).))))))...))))))))..)))))..((((((........(((....((((.....))))..)))))))-------)) ( -30.20) >DroSec_CAF1 13672 113 + 1 AAACUAUUUUCGCCUGUGGACAAUAAGCGAGCAAGUGCCGCAAUUGCUGACCAGGCGAACUAAUAGAUGCGAUUUUAAAUGUGCUCUCAAGAUUCUAUAUCUGCAGCAUUC-------GU ...(((((((((((((.((.((((..(((.((....))))).))))))...))))))))..)))))..((((........(((((....((((.....))))..)))))))-------)) ( -31.70) >DroSim_CAF1 24921 113 + 1 AAACUAUUUUCGCCUGUGGACAAUAAGCGAGCAAGUGCCGCAAUUGCUGACCAGGCGAACUAAUAGAUGCGAAUUUAAAUGUGCUCUCAAGAUUCUAUAUCUGCAGCAUUC-------GU ...(((((((((((((.((.((((..(((.((....))))).))))))...))))))))..)))))..((((........(((((....((((.....))))..)))))))-------)) ( -31.70) >DroEre_CAF1 13426 110 + 1 AAGCUAUUUUCGCCUGUGGACAAUGAGCGAGCCAGUGCCGCAAUUGCUA----GUGCCUAUAGUAGAUGCGAAUUUAAAUGUGC--UCAAGAUUCU----CUGCAGCAUUCUAACGUGCA ..((.......((.((..((.((((((((.((....))((((.((((((----.......)))))).))))..........)))--)))...)).)----)..))))...(....).)). ( -25.20) >DroYak_CAF1 13677 113 + 1 AAACUAUUUUCGCUUGCGGACAAUAAGCGAGCGAGUGCCGCAACUGCUGAGCAGGCGGAAUAAUAGAUGCGAAUUUAAAUGUGCUCUCAAGAUUCUAUAUCUGCGGCAUUC-------CU ...........((((((.........))))))((((((((((.((((...))))..(((((...(((.(((.((....)).))))))....))))).....))))))))))-------.. ( -36.80) >consensus AAACUAUUUUCGCCUGUGGACAAUAAGCGAGCAAGUGCCGCAAUUGCUGACCAGGCGAACUAAUAGAUGCGAAUUUAAAUGUGCUCUCAAGAUUCUAUAUCUGCAGCAUUC_______GU ...((((((((((((((((.((((..(((.((....))))).)))))))..))))))))..)))))..............(((((....((((.....))))..)))))........... (-22.52 = -24.00 + 1.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:47 2006