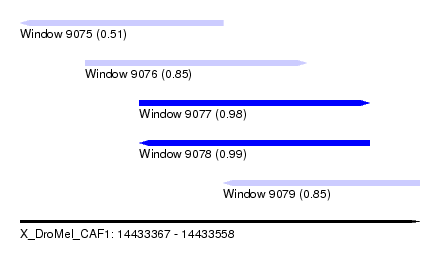
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,433,367 – 14,433,558 |
| Length | 191 |
| Max. P | 0.991287 |

| Location | 14,433,367 – 14,433,464 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -32.06 |
| Consensus MFE | -27.42 |
| Energy contribution | -27.86 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14433367 97 - 22224390 GUGCGCCGGCGCUGCGAGAGUCGAGCUUUUCCCGCAGAAUACCGGUUUCCCAU--------------UUCCCGACUUUUGGCUAUUUUUGGCCCAGAGUCGCUAUAC--AUAU ..(((((((..(((((.(((........))).)))))....))))).......--------------.....((((((.(((((....))))).)))))))).....--.... ( -34.50) >DroSec_CAF1 12712 97 - 1 GUGCGCCGGCGCUGCGAGAGCCGAGCUUUUACCGCAGAAUACCGGUUUCCCAU--------------UUCCCGACUUUUGGCUAUUUUUGGCCCAGAGUCGCUGUAU--AUAU (((((((((..((((((((((...)))))...)))))....))))).......--------------....(((((((.(((((....))))).)))))))..))))--.... ( -35.60) >DroSim_CAF1 23935 97 - 1 GUGCGCCGGCGCUGCGAGAGCCGAGCUUUUACCACAGAAUACCGGUUUCCCAU--------------UUCCCGACUUUUGGCUAUUUUUGGCCCAGAGUCGCUGUAC--AUAU (((((((((..(((.((((((...))))))....)))....))))).......--------------....(((((((.(((((....))))).)))))))..))))--.... ( -33.10) >DroEre_CAF1 12434 110 - 1 GAGCGCCGGCGCCGCGAGAGCCGAGCUUUC-CUGCAGAAUACCGGUUUCCCGUUUCCCAUUUCCCAUUUCCCGACUUU--GCUAUUUUUGGCCCAGAGUCGAGCCACACACAU ((((..((((..(....).)))).))))..-..((.(((...(((....))).)))...............(((((((--((((....))))..))))))).))......... ( -28.70) >DroYak_CAF1 12576 98 - 1 GAGCGCCGGCGCUGGGAGAGUCGAGCUUUU-CCGCAGAAUACUGGUUUCCCAUU-------------UUCCCGACUUU-GGCUAUUUUUGGCCCAGACUCGAGCCACACAAAU .......(((((.(((((((.....)))))-)))).(((...(((....)))..-------------))).(((.(((-(((((....))).))))).))).)))........ ( -28.40) >consensus GUGCGCCGGCGCUGCGAGAGCCGAGCUUUU_CCGCAGAAUACCGGUUUCCCAU______________UUCCCGACUUUUGGCUAUUUUUGGCCCAGAGUCGCUCUAC__AUAU ....(((((..((((((((((...))))))...))))....))))).........................(((((((.(((((....))))).)))))))............ (-27.42 = -27.86 + 0.44)



| Location | 14,433,398 – 14,433,504 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.65 |
| Mean single sequence MFE | -41.62 |
| Consensus MFE | -39.04 |
| Energy contribution | -38.28 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14433398 106 + 22224390 CAAAAGUCGGGAA--------------AUGGGAAACCGGUAUUCUGCGGGAAAAGCUCGACUCUCGCAGCGCCGGCGCACUGAAUGAUGUCCUUGCCAGGACAUCUCGCUGCCAUUGCUG .....(((((...--------------.(((....))).....((((((((..........))))))))..)))))(((.(((..((((((((....))))))))))).)))........ ( -39.60) >DroSec_CAF1 12743 106 + 1 CAAAAGUCGGGAA--------------AUGGGAAACCGGUAUUCUGCGGUAAAAGCUCGGCUCUCGCAGCGCCGGCGCACUGAAUGAUGUCCUUGCCAGGACAUCUCGCUGCCGUCGCUG ....(((((((((--------------.(((....)))...))))((.......)).)))))....(((((.((((((...((..((((((((....)))))))))))).)))).))))) ( -40.50) >DroSim_CAF1 23966 106 + 1 CAAAAGUCGGGAA--------------AUGGGAAACCGGUAUUCUGUGGUAAAAGCUCGGCUCUCGCAGCGCCGGCGCACUGUAUGAUGUCCUUGCCAGGACAUCUCGCUGCCGUCGCUG ....(((((((..--------------.(((....))).(((......)))....)))))))....(((((.(((((....((..((((((((....))))))))..))))))).))))) ( -39.80) >DroEre_CAF1 12467 117 + 1 --AAAGUCGGGAAAUGGGAAAUGGGAAACGGGAAACCGGUAUUCUGCAG-GAAAGCUCGGCUCUCGCGGCGCCGGCGCUCUGGACGAUGUCCUUGCCAGGACAUCUCGCUGCCGUCGCUG --.....(((((..((((...((((((.(((....)))...)))).)).-.....))))..)))))(((((.((((((....((.((((((((....)))))))))))).)))).))))) ( -45.90) >DroYak_CAF1 12609 105 + 1 C-AAAGUCGGGAA-------------AAUGGGAAACCAGUAUUCUGCGG-AAAAGCUCGACUCUCCCAGCGCCGGCGCUCUGGAUGAUGUCCUUGCCAGGACAUCUCGCUGCCGUUGCUG .-..(((((((((-------------..(((....)))...))))((..-....)).)))))....(((((.((((((....((.((((((((....)))))))))))).)))).))))) ( -42.30) >consensus CAAAAGUCGGGAA______________AUGGGAAACCGGUAUUCUGCGG_AAAAGCUCGGCUCUCGCAGCGCCGGCGCACUGAAUGAUGUCCUUGCCAGGACAUCUCGCUGCCGUCGCUG ....(((((((.................(((....))).....((........)))))))))....(((((.((((((.......((((((((....))))))))..)).)))).))))) (-39.04 = -38.28 + -0.76)


| Location | 14,433,424 – 14,433,534 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -43.60 |
| Consensus MFE | -36.90 |
| Energy contribution | -36.98 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14433424 110 + 22224390 AUUCUGCGGGAAAAGCUCGACUCUCGCAGCGCCGGCGCACUGAAUGAUGUCCUUGCCAGGACAUCUCGCUGCCAUUGCUGAUAACAAUCCGCAGGCUGUGUUAUCGCCGC ...((((((((..........))))))))...(((((........((((((((....)))))))).(((.(((..(((.(((....))).)))))).)))....))))). ( -41.00) >DroSec_CAF1 12769 110 + 1 AUUCUGCGGUAAAAGCUCGGCUCUCGCAGCGCCGGCGCACUGAAUGAUGUCCUUGCCAGGACAUCUCGCUGCCGUCGCUGAUAACAAUUCGCAGGCUGUGUUAUCGCCGC .....((((..(.(((.((((.((.((((((.((((((...((..((((((((....)))))))))))).)))).))))((.......)))))))))).))).)..)))) ( -44.30) >DroSim_CAF1 23992 110 + 1 AUUCUGUGGUAAAAGCUCGGCUCUCGCAGCGCCGGCGCACUGUAUGAUGUCCUUGCCAGGACAUCUCGCUGCCGUCGCUGAUAACAAUUCGCAGGCUGUGUUAUCGCCGC .....((((..(.(((.((((.((.((((((.(((((....((..((((((((....))))))))..))))))).))))((.......)))))))))).))).)..)))) ( -43.10) >DroEre_CAF1 12505 109 + 1 AUUCUGCAG-GAAAGCUCGGCUCUCGCGGCGCCGGCGCUCUGGACGAUGUCCUUGCCAGGACAUCUCGCUGCCGUCGCUGAUAGCAAUUCGCAGGCGGCGUUAUCGCCGC ...((((.(-((..(((((((....((((((((((....))))..((((((((....)))))))).))))))....)))))..))..)))))))((((((....)))))) ( -49.50) >DroYak_CAF1 12635 109 + 1 AUUCUGCGG-AAAAGCUCGACUCUCCCAGCGCCGGCGCUCUGGAUGAUGUCCUUGCCAGGACAUCUCGCUGCCGUUGCUGAUAACAAUUCGCAGGCUGCGUUAUCGCCGC ...((((((-(.......(.....).(((((.((((((....((.((((((((....)))))))))))).)))).))))).......)))))))((.(((....))).)) ( -40.10) >consensus AUUCUGCGG_AAAAGCUCGGCUCUCGCAGCGCCGGCGCACUGAAUGAUGUCCUUGCCAGGACAUCUCGCUGCCGUCGCUGAUAACAAUUCGCAGGCUGUGUUAUCGCCGC ...((((((.......(((((....((((((.(((....)))...((((((((....)))))))).))))))....))))).......))))))((.(((....))).)) (-36.90 = -36.98 + 0.08)


| Location | 14,433,424 – 14,433,534 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -45.40 |
| Consensus MFE | -38.04 |
| Energy contribution | -38.24 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14433424 110 - 22224390 GCGGCGAUAACACAGCCUGCGGAUUGUUAUCAGCAAUGGCAGCGAGAUGUCCUGGCAAGGACAUCAUUCAGUGCGCCGGCGCUGCGAGAGUCGAGCUUUUCCCGCAGAAU ((((.((.......(((.(((.((((((..((....))..)))..((((((((....))))))))....))).))).)))(((.((.....)))))...))))))..... ( -43.60) >DroSec_CAF1 12769 110 - 1 GCGGCGAUAACACAGCCUGCGAAUUGUUAUCAGCGACGGCAGCGAGAUGUCCUGGCAAGGACAUCAUUCAGUGCGCCGGCGCUGCGAGAGCCGAGCUUUUACCGCAGAAU ((((.(((((((..(....)....)))))))(((..((((..((.((((((((....))))))))...((((((....))))))))...)))).)))....))))..... ( -45.80) >DroSim_CAF1 23992 110 - 1 GCGGCGAUAACACAGCCUGCGAAUUGUUAUCAGCGACGGCAGCGAGAUGUCCUGGCAAGGACAUCAUACAGUGCGCCGGCGCUGCGAGAGCCGAGCUUUUACCACAGAAU .(((((((((((..(....)....)))))))((((.((((.(((.((((((((....))))))))......))))))).))))......))))................. ( -41.50) >DroEre_CAF1 12505 109 - 1 GCGGCGAUAACGCCGCCUGCGAAUUGCUAUCAGCGACGGCAGCGAGAUGUCCUGGCAAGGACAUCGUCCAGAGCGCCGGCGCCGCGAGAGCCGAGCUUUC-CUGCAGAAU ((((((....))))))(((((..(((((..(......)..)))))((((((((....))))))))....(((((..((((..(....).)))).))))).-.)))))... ( -50.10) >DroYak_CAF1 12635 109 - 1 GCGGCGAUAACGCAGCCUGCGAAUUGUUAUCAGCAACGGCAGCGAGAUGUCCUGGCAAGGACAUCAUCCAGAGCGCCGGCGCUGGGAGAGUCGAGCUUUU-CCGCAGAAU ((.(((....))).))(((((.........((((..((((.((((((((((((....)))))))).))....))))))..))))((((((.....)))))-))))))... ( -46.00) >consensus GCGGCGAUAACACAGCCUGCGAAUUGUUAUCAGCGACGGCAGCGAGAUGUCCUGGCAAGGACAUCAUCCAGUGCGCCGGCGCUGCGAGAGCCGAGCUUUU_CCGCAGAAU ((((.(((((((..(....)....)))))))(((..((((.....((((((((....))))))))...((((((....)))))).....)))).)))....))))..... (-38.04 = -38.24 + 0.20)


| Location | 14,433,464 – 14,433,558 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -34.34 |
| Consensus MFE | -26.58 |
| Energy contribution | -26.62 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14433464 94 - 22224390 GCGUUG-UGGCAGUGAAAGUUUUGUGCGGCGAUAACACAGCCUGCGGAUUGUUAUCAGCAAUGGCAGCGAGAUGUCCUGGCAAGGACAUCAUUCA ((((((-(.((.((...........)).)).)))))...((((((.(((....))).)))..))).))..((((((((....))))))))..... ( -32.90) >DroSec_CAF1 12809 94 - 1 GCGGUGUUGGCAGUGAAAGU-UGGUGCGGCGAUAACACAGCCUGCGAAUUGUUAUCAGCGACGGCAGCGAGAUGUCCUGGCAAGGACAUCAUUCA ((.(..(..((.......))-..)..).))(((((((..(....)....))))))).((.......))..((((((((....))))))))..... ( -32.10) >DroSim_CAF1 24032 94 - 1 GCGGUGUUGGCAGUGAAAGU-UUGUGCGGCGAUAACACAGCCUGCGAAUUGUUAUCAGCGACGGCAGCGAGAUGUCCUGGCAAGGACAUCAUACA .((.(((((((.((((.(((-(((((.(((.........))))))))))).))))..))..))))).)).((((((((....))))))))..... ( -32.80) >DroEre_CAF1 12544 93 - 1 GCGUGG-UGGCAGUGAAAGU-UUUUGCGGCGAUAACGCCGCCUGCGAAUUGCUAUCAGCGACGGCAGCGAGAUGUCCUGGCAAGGACAUCGUCCA ((.(((-(((((((....((-....((((((....))))))..))..))))))))))))((((....)..((((((((....))))))))))).. ( -41.60) >DroYak_CAF1 12674 93 - 1 GCGGUG-UGGCAGUGAAAGU-UUUUGCGGCGAUAACGCAGCCUGCGAAUUGUUAUCAGCAACGGCAGCGAGAUGUCCUGGCAAGGACAUCAUCCA (((.((-(.((.(..((...-.))..).)).))).))).((((((..((....))..)))..))).(.((((((((((....)))))))).))). ( -32.30) >consensus GCGGUG_UGGCAGUGAAAGU_UUGUGCGGCGAUAACACAGCCUGCGAAUUGUUAUCAGCGACGGCAGCGAGAUGUCCUGGCAAGGACAUCAUCCA ((..((.(((((((..........((((((.........))).))).))))))).))((....)).))..((((((((....))))))))..... (-26.58 = -26.62 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:44 2006