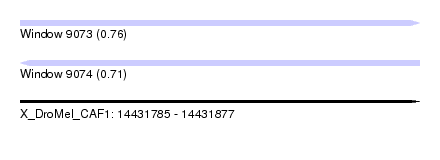
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,431,785 – 14,431,877 |
| Length | 92 |
| Max. P | 0.756909 |

| Location | 14,431,785 – 14,431,877 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 84.46 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -20.20 |
| Energy contribution | -21.15 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
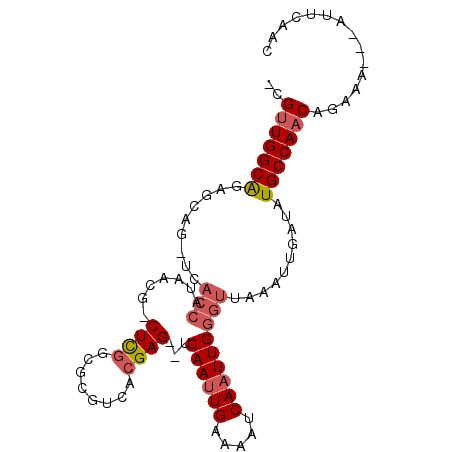
>X_DroMel_CAF1 14431785 92 + 22224390 GUUGAAU---UUUCCGUUGGCAUAUCAAUUUAACCCAAUUGAUUUUCAAUUGA--CUCGUGACGCGCCGAG-CGUUAUGGUGCUCUGCUCUGCCAACG- .......---....((((((((.............((((((.....))))))(--(.((((((((.....)-))))))))).........))))))))- ( -29.30) >DroPse_CAF1 12168 85 + 1 UCUGUAUCUGUAUCUGUUGGCAUUUCAAUUUAACUCAAUUGAUUUUCAUUUGAAUCUCGUGACGCGCUAAG-CC-------------CUCUGCCAGCGC ...............(((((((............(((...(((((......)))))...))).((.....)-).-------------...))))))).. ( -17.20) >DroSec_CAF1 11127 92 + 1 GUUGAAU---UUUCCGUUGGCAUAUCGAUUUAACCCAAUUGAUUUUCAAUUGA--CUCGUGACGCUCCGAGCCGUUAUGGUGA-CUGCUCUGCCAACG- .......---....((((((((.............((((((.....))))))(--(.((((((((.....).)))))))))..-......))))))))- ( -25.00) >DroSim_CAF1 22377 92 + 1 GUUGAAU---UUUCCGUUGGCAUAUCAAUUUAACCCAAUUGAUUUUCAAUUGA--CUCGUGACGCGCCGAGCCGUUAUGGUGA-CUGCUCUGCCAACG- .......---....((((((((.............((((((.....))))))(--(.(((((((.((...)))))))))))..-......))))))))- ( -26.50) >DroEre_CAF1 10917 91 + 1 GUCGUCU---CUUCUGUUGGCAUAUCAAUUUAACCCAAUUGAUUUUCAAUUGA--CUCGUGACGCGCCGAG-CGUUAUGGUGA-CCGCUCCGCCAACG- .......---.....((((((..............((((((.....))))))(--(.((((((((.....)-)))))))))..-.......)))))).- ( -28.00) >DroYak_CAF1 10994 91 + 1 GUUGCCU---UUUCUGUUGGCAUAUUAAUUUAACCCAAUUGAUUUUCAAUUGA--CUCGUGACGCGCCGAG-CGUUAUGGUGA-CUGCUCUGCCAACG- .......---.....(((((((.............((((((.....))))))(--(.((((((((.....)-)))))))))..-......))))))).- ( -28.30) >consensus GUUGAAU___UUUCCGUUGGCAUAUCAAUUUAACCCAAUUGAUUUUCAAUUGA__CUCGUGACGCGCCGAG_CGUUAUGGUGA_CUGCUCUGCCAACG_ ..............(((((((..............((((((.....))))))....(((((((((.....).))))))))...........))))))). (-20.20 = -21.15 + 0.95)



| Location | 14,431,785 – 14,431,877 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 84.46 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -19.54 |
| Energy contribution | -20.26 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14431785 92 - 22224390 -CGUUGGCAGAGCAGAGCACCAUAACG-CUCGGCGCGUCACGAG--UCAAUUGAAAAUCAAUUGGGUUAAAUUGAUAUGCCAACGGAAA---AUUCAAC -(((((((((.((.((((........)-))).)).)((((.((.--(((((((.....))))))).))....)))).))))))))....---....... ( -31.00) >DroPse_CAF1 12168 85 - 1 GCGCUGGCAGAG-------------GG-CUUAGCGCGUCACGAGAUUCAAAUGAAAAUCAAUUGAGUUAAAUUGAAAUGCCAACAGAUACAGAUACAGA ((((((((....-------------.)-).))))))(((....(((((((.((.....)).)))))))...(((......)))..)))........... ( -18.60) >DroSec_CAF1 11127 92 - 1 -CGUUGGCAGAGCAG-UCACCAUAACGGCUCGGAGCGUCACGAG--UCAAUUGAAAAUCAAUUGGGUUAAAUCGAUAUGCCAACGGAAA---AUUCAAC -((((((((((((.(-(.......)).))))....((....((.--(((((((.....))))))).))....))...))))))))....---....... ( -27.80) >DroSim_CAF1 22377 92 - 1 -CGUUGGCAGAGCAG-UCACCAUAACGGCUCGGCGCGUCACGAG--UCAAUUGAAAAUCAAUUGGGUUAAAUUGAUAUGCCAACGGAAA---AUUCAAC -((((((((...(((-(.(((.....((((((........))))--))(((((.....))))).)))...))))...))))))))....---....... ( -31.50) >DroEre_CAF1 10917 91 - 1 -CGUUGGCGGAGCGG-UCACCAUAACG-CUCGGCGCGUCACGAG--UCAAUUGAAAAUCAAUUGGGUUAAAUUGAUAUGCCAACAGAAG---AGACGAC -.(((((((...(((-(.(((.....(-((((........))))--)((((((.....)))))))))...))))...))))))).....---....... ( -27.70) >DroYak_CAF1 10994 91 - 1 -CGUUGGCAGAGCAG-UCACCAUAACG-CUCGGCGCGUCACGAG--UCAAUUGAAAAUCAAUUGGGUUAAAUUAAUAUGCCAACAGAAA---AGGCAAC -.(((((((......-..(((.....(-((((........))))--)((((((.....)))))))))..........))))))).....---....... ( -25.27) >consensus _CGUUGGCAGAGCAG_UCACCAUAACG_CUCGGCGCGUCACGAG__UCAAUUGAAAAUCAAUUGGGUUAAAUUGAUAUGCCAACAGAAA___AUUCAAC ..(((((((.........(((.......((((........))))...((((((.....)))))))))..........)))))))............... (-19.54 = -20.26 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:40 2006