
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,430,374 – 14,430,485 |
| Length | 111 |
| Max. P | 0.979807 |

| Location | 14,430,374 – 14,430,485 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.26 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -13.18 |
| Energy contribution | -13.02 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14430374 111 + 22224390 UUUAUUGCUACUCAAAAAAAAAAAAUGGUGCAAACCGGUUAUAAACCGAUUUUGAGUGGCUUGUGUGGUUAUUUAAUAACACCAAAUAAAUUGAUAAUAAAU---------AAUUACAUA ......(((((((((((.........(((....)))(((.....)))..))))))))))).((((((((((((((.((.((..........)).)).)))))---------))))))))) ( -30.40) >DroSec_CAF1 9699 113 + 1 UUUAUUGCCACUCAAAUUGCAAA-------UUAACCGGUUAUAAGCCGAUUUUGAGUGGUUAGUGUGGUUAAUUAACAAUACCAAAUAGUGUGGUGAUGUUUUGAUACUUUUGUUAUAUA ..((((((((((((((.......-------.....((((.....))))..))))))))).))))).(((.....((((.(((((.......))))).)))).....)))........... ( -24.44) >DroSim_CAF1 21010 113 + 1 UUUAUUGCCACUCAAAUUGCAAA-------UUAACCGGUUAUAAGCCGAUUUUGAGUGGUUAGUGUGGUUAAUUAACAAUACCAAAUAGUGUGGUGAUGUUUUGAUACUUUUGUUAUAUA ..((((((((((((((.......-------.....((((.....))))..))))))))).))))).(((.....((((.(((((.......))))).)))).....)))........... ( -24.44) >DroEre_CAF1 9406 113 + 1 UGUAUUGCUACUGAGACCAAU-------UGCAAACCUGUUAUAAGCCGAUUUCGAGUGGUUCGUGUGGUUAAUUUAUAAUACCAAAUAAUGUGGUGAUGAUUUGAUACUAUUGCUAUAUA .((((((((((...(((((.(-------((.(((.(.((.....)).).)))))).)))))...)))))........)))))......((((((..(((.........)))..)))))). ( -19.90) >DroYak_CAF1 9591 108 + 1 UGUAUUGCCGCUAAAACCACU-------UGCAAACCGGUUAUAAGCCGAUUUCGAGUGGUUAGUGUGGUUAAAUAAUAACACCAAAUAAUUCGCUAUAGAUUUGAUACU-----UAUAUA .((((((((((...(((((((-------((.(((.((((.....)))).))))))))))))...))))).....................((......))...))))).-----...... ( -27.90) >consensus UUUAUUGCCACUCAAACCACAAA_____UGCAAACCGGUUAUAAGCCGAUUUUGAGUGGUUAGUGUGGUUAAUUAAUAAUACCAAAUAAUGUGGUGAUGAUUUGAUACU_UUGUUAUAUA ......((((((((((...................((((.....))))..))))))))))..((((.(((....))).))))...................................... (-13.18 = -13.02 + -0.16)

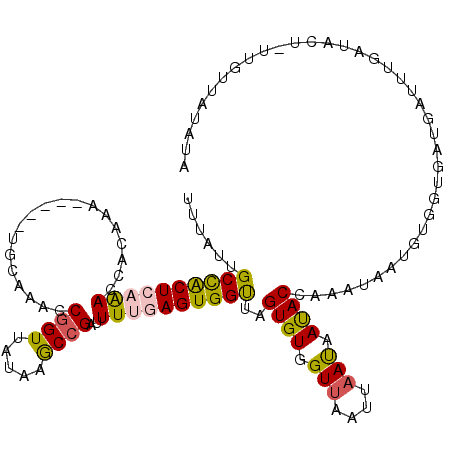

| Location | 14,430,374 – 14,430,485 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.26 |
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -9.16 |
| Energy contribution | -10.32 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14430374 111 - 22224390 UAUGUAAUU---------AUUUAUUAUCAAUUUAUUUGGUGUUAUUAAAUAACCACACAAGCCACUCAAAAUCGGUUUAUAACCGGUUUGCACCAUUUUUUUUUUUUGAGUAGCAAUAAA ..(((..((---------(((((.((((((.....))))))....)))))))..)))...((.((((((((..(((.....)))(((....))).........)))))))).))...... ( -23.20) >DroSec_CAF1 9699 113 - 1 UAUAUAACAAAAGUAUCAAAACAUCACCACACUAUUUGGUAUUGUUAAUUAACCACACUAACCACUCAAAAUCGGCUUAUAACCGGUUAA-------UUUGCAAUUUGAGUGGCAAUAAA ...................((((..((((.......))))..))))...............((((((((((((((.......)))))...-------.......)))))))))....... ( -20.70) >DroSim_CAF1 21010 113 - 1 UAUAUAACAAAAGUAUCAAAACAUCACCACACUAUUUGGUAUUGUUAAUUAACCACACUAACCACUCAAAAUCGGCUUAUAACCGGUUAA-------UUUGCAAUUUGAGUGGCAAUAAA ...................((((..((((.......))))..))))...............((((((((((((((.......)))))...-------.......)))))))))....... ( -20.70) >DroEre_CAF1 9406 113 - 1 UAUAUAGCAAUAGUAUCAAAUCAUCACCACAUUAUUUGGUAUUAUAAAUUAACCACACGAACCACUCGAAAUCGGCUUAUAACAGGUUUGCA-------AUUGGUCUCAGUAGCAAUACA ((((.(((.((((((((((((.((......)).))))))))))))............(((.....)))......)))))))...(..((((.-------((((....)))).))))..). ( -19.30) >DroYak_CAF1 9591 108 - 1 UAUAUA-----AGUAUCAAAUCUAUAGCGAAUUAUUUGGUGUUAUUAUUUAACCACACUAACCACUCGAAAUCGGCUUAUAACCGGUUUGCA-------AGUGGUUUUAGCGGCAAUACA ......-----...............((........((((...........))))(.(((((((((.((((((((.......))))))).).-------))))))..))).)))...... ( -21.80) >consensus UAUAUAACAA_AGUAUCAAAUCAUCACCACAUUAUUUGGUAUUAUUAAUUAACCACACUAACCACUCAAAAUCGGCUUAUAACCGGUUUGCA_____UUUGUAAUUUGAGUGGCAAUAAA ....................................((((...........))))......((((((((((((((.......)))))).................))))))))....... ( -9.16 = -10.32 + 1.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:37 2006