
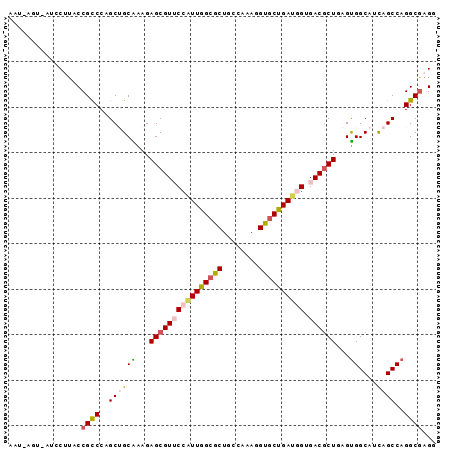
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,421,189 – 14,421,320 |
| Length | 131 |
| Max. P | 0.766029 |

| Location | 14,421,189 – 14,421,280 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 82.85 |
| Mean single sequence MFE | -34.03 |
| Consensus MFE | -27.41 |
| Energy contribution | -27.67 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14421189 91 + 22224390 AGUCA---AUCCUUACCGCCCAGCUGCGAAGAGCGUUCCAUUGGCGCUGCCAAAGGUGCUGAUGGUGACGCUGAGUGGCAUCAGCCAGGCGAGC .....---........((((((((.((.....))(((((((..(((((......)))))..)))).)))))))..((((....))))))))... ( -37.70) >DroGri_CAF1 1462 92 + 1 AAUAUGU--AACGUACCGUCCUGCGGCAAAGAGCGUACCGUUAGCACUACCAAAUGUACUGAUAGUGACGCUCAGUGGCAUUAGCCAGGCGAGG .......--..........(((.((.(...((((((((..((((.((........)).))))..)).))))))..((((....))))).))))) ( -27.60) >DroSim_CAF1 864 94 + 1 AGUCAAUAAUCCUUACCGUCCAGCUGCGAAGAGCGUUCCAUUGGCGCUGCCAAAGGUGCUGAUGGUGACGCUGAGUGGCAUCAGCCAGGCGAGC ...........((..((...((((.((.....))(((((((..(((((......)))))..)))).)))))))..((((....))))))..)). ( -35.10) >DroEre_CAF1 931 93 + 1 AGU-AGUGAUCCUUACCGCCCAGCUGCGAAGAGCGUUCCAUUGGCGCUGCCAAAGGUGCUGAUGGUGACGCUCAGCGGCAUCAGCCAGGCGAGG ...-......(((..((((...(((((...(((((((((((..(((((......)))))..)))).))))))).)))))....))..))..))) ( -42.60) >DroYak_CAF1 899 93 + 1 AAU-AAUAAUCCUUACCGCCCAGCUGCAAAGAGCGUUCCAUUGGCGCUGCCAAACGUGCUGAUGGUGACGCUAAGUGGCAUCAGCCAGGCGAGU ...-............((((..((((((...((((((((((..((((........))))..)))).))))))...))....))))..))))... ( -36.50) >DroMoj_CAF1 410 92 + 1 CAUUUGU--AACAUACCGUCCUGCGGCAAAGAGCGUACCAUUAGCACUUCCAAAUGUGCUGACAGUGACACUGAGUGGCAUUAGCCAGGCAAGG .......--......((((((((..((.....))......(((((((........)))))))))).)))..((..((((....))))..)).)) ( -24.70) >consensus AAU_AGU_AUCCUUACCGCCCAGCUGCAAAGAGCGUUCCAUUGGCGCUGCCAAAGGUGCUGAUGGUGACGCUGAGUGGCAUCAGCCAGGCGAGG ................((((..((((((...((((((((((((((((........)))))))))).))))))...))....))))..))))... (-27.41 = -27.67 + 0.25)


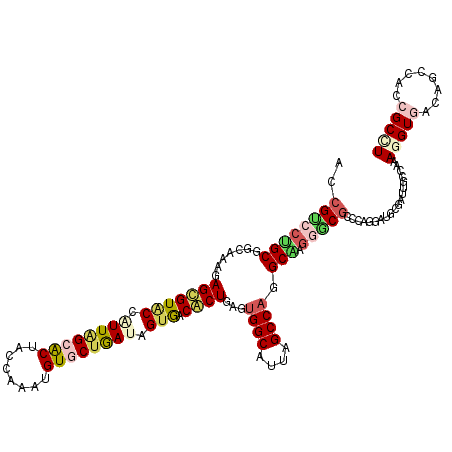
| Location | 14,421,200 – 14,421,320 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.06 |
| Mean single sequence MFE | -45.17 |
| Consensus MFE | -26.33 |
| Energy contribution | -27.37 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14421200 120 + 22224390 ACCGCCCAGCUGCGAAGAGCGUUCCAUUGGCGCUGCCAAAGGUGCUGAUGGUGACGCUGAGUGGCAUCAGCCAGGCGAGCGCGCCCAGGAUGCGAUUGCCGAAGGUGACGGCCACCGCCU ...........(((...((((((((((..(((((......)))))..)))).))))))..(((((....(((.(((((.(((((....).)))).)))))...)))....)))))))).. ( -54.80) >DroVir_CAF1 14702 120 + 1 ACCGUCCUGCGGCAAAGAGUGUACCAUUAGCACUACCAAAUGUGCUAACAGUAACACUAAGUGGCAUUAGCCAAGCAAGGGCGCCUAAAAUACGAUUGCCAAAGGUGACAGCGACCGCCU ..((((((...((....(((((((..(((((((........)))))))..)).)))))...((((....)))).)).))))))...................(((((........))))) ( -37.80) >DroGri_CAF1 1474 120 + 1 ACCGUCCUGCGGCAAAGAGCGUACCGUUAGCACUACCAAAUGUACUGAUAGUGACGCUCAGUGGCAUUAGCCAGGCGAGGACGCCCAGGAUGCGAUUGCCAAAGGUGACAGCGACCGCCU ...((((((.(((...((((((((..((((.((........)).))))..)).))))))..((((....)))).........)))))))))(((.((((....(....).)))).))).. ( -45.20) >DroWil_CAF1 1056 120 + 1 ACCGUCCGGCAGCAAAGAGUGUACCAUUUGCACUACCAAAUGUACUAAUGGUAACACUCAAUGGCAUUAGCCAGGCCAAGGCGCCCAAUAUCCGAUUGCCAAAGGUUACCGCCACCGCAU (((....(((......((((((((((((.(.((........)).).)))))).))))))..((((....)))).)))..((((..(.......)..))))...)))....((....)).. ( -34.70) >DroMoj_CAF1 422 120 + 1 ACCGUCCUGCGGCAAAGAGCGUACCAUUAGCACUUCCAAAUGUGCUGACAGUGACACUGAGUGGCAUUAGCCAGGCAAGGGCGCCAAGGAUGCGAUUGCCAAAGGUCACAGCAACCGCUU .(((.....)))....(((((.....(((((((........)))))))..(((((.((...((((....))))(((((..((((....).)))..)))))..)))))))......))))) ( -42.70) >DroAna_CAF1 2318 120 + 1 ACCGGCCGGCAGCGAAGAGCGUGCCAUUGGCACUGCCAAAGGUGCUGAUGGUGACGCUGAGGGGCAUGAGCCACGCCAGCGCUCCGAGGACGCGGUUCCCAAAAGUGACCGCCACCGCCU ...((((((.((((...((((((((((..(((((......)))))..))))).)))))..(((((....)))...))..))))))).((..(((((..(.....)..)))))..))))). ( -55.80) >consensus ACCGUCCUGCGGCAAAGAGCGUACCAUUAGCACUACCAAAUGUGCUGAUAGUGACACUGAGUGGCAUUAGCCAGGCAAGGGCGCCCAGGAUGCGAUUGCCAAAGGUGACAGCCACCGCCU ..((((((((.......(((((((.((((((((........)))))))).))).))))...((((....)))).))).)))))...................(((((........))))) (-26.33 = -27.37 + 1.03)

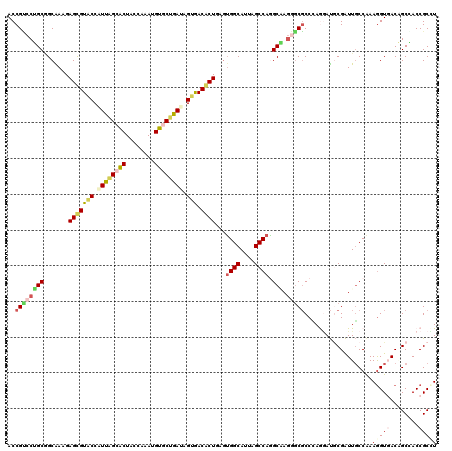
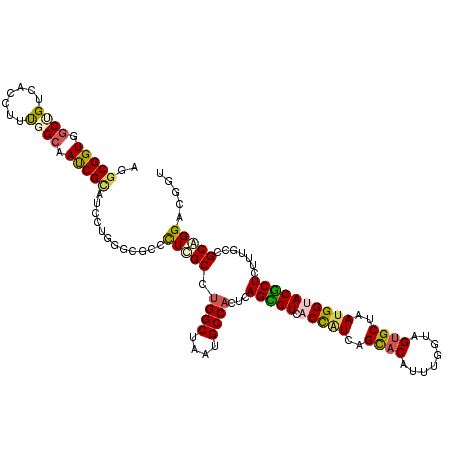
| Location | 14,421,200 – 14,421,320 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.06 |
| Mean single sequence MFE | -47.17 |
| Consensus MFE | -31.37 |
| Energy contribution | -31.27 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14421200 120 - 22224390 AGGCGGUGGCCGUCACCUUCGGCAAUCGCAUCCUGGGCGCGCUCGCCUGGCUGAUGCCACUCAGCGUCACCAUCAGCACCUUUGGCAGCGCCAAUGGAACGCUCUUCGCAGCUGGGCGGU .((((.((((((.......))))....((((((..((((....))))..)..)))))....)).))))(((.(((((.((.(((((...))))).))...((.....)).)))))..))) ( -49.40) >DroVir_CAF1 14702 120 - 1 AGGCGGUCGCUGUCACCUUUGGCAAUCGUAUUUUAGGCGCCCUUGCUUGGCUAAUGCCACUUAGUGUUACUGUUAGCACAUUUGGUAGUGCUAAUGGUACACUCUUUGCCGCAGGACGGU .(((((...))))).(((.((((((.........(((((....)))))(((....)))....(((((.(((((((((((........))))))))))))))))..)))))).)))..... ( -41.90) >DroGri_CAF1 1474 120 - 1 AGGCGGUCGCUGUCACCUUUGGCAAUCGCAUCCUGGGCGUCCUCGCCUGGCUAAUGCCACUGAGCGUCACUAUCAGUACAUUUGGUAGUGCUAACGGUACGCUCUUUGCCGCAGGACGGU .(((((...))))).(((.((((((..((((.(..((((....))))..)...))))....(((((((((((((((.....)))))))))(....)..)))))).)))))).)))..... ( -46.80) >DroWil_CAF1 1056 120 - 1 AUGCGGUGGCGGUAACCUUUGGCAAUCGGAUAUUGGGCGCCUUGGCCUGGCUAAUGCCAUUGAGUGUUACCAUUAGUACAUUUGGUAGUGCAAAUGGUACACUCUUUGCUGCCGGACGGU ...((.(((((((((....(((((.(..(...(..(((......)))..))..))))))..((((((.((((((.((((........)))).)))))))))))).)))))))))..)).. ( -46.00) >DroMoj_CAF1 422 120 - 1 AAGCGGUUGCUGUGACCUUUGGCAAUCGCAUCCUUGGCGCCCUUGCCUGGCUAAUGCCACUCAGUGUCACUGUCAGCACAUUUGGAAGUGCUAAUGGUACGCUCUUUGCCGCAGGACGGU ..((((((((((.......)))))))))).((((((((.....((..((((....))))..))((((.(((((.(((((........))))).))))))))).....)))).)))).... ( -45.40) >DroAna_CAF1 2318 120 - 1 AGGCGGUGGCGGUCACUUUUGGGAACCGCGUCCUCGGAGCGCUGGCGUGGCUCAUGCCCCUCAGCGUCACCAUCAGCACCUUUGGCAGUGCCAAUGGCACGCUCUUCGCUGCCGGCCGGU .((((((((((((..(.....)..)))))......((.(((((((.(.(((....)))).)))))))..)).....))))..((((((((((...)))))((.....))))))))))... ( -53.50) >consensus AGGCGGUGGCUGUCACCUUUGGCAAUCGCAUCCUGGGCGCCCUCGCCUGGCUAAUGCCACUCAGCGUCACCAUCAGCACAUUUGGUAGUGCUAAUGGUACGCUCUUUGCCGCAGGACGGU ..(((((.((((.......)))).)))))............(((((.((((....))))...(((((.(((((..((((........))))..)))))))))).......)))))..... (-31.37 = -31.27 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:26 2006