| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,417,919 – 14,418,039 |
| Length | 120 |
| Max. P | 0.510974 |

| Location | 14,417,919 – 14,418,039 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.15 |
| Mean single sequence MFE | -37.23 |
| Consensus MFE | -22.78 |
| Energy contribution | -23.28 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14417919 120 - 22224390 UGGUGGGACCCAGAAGACUGCGCCACUUCUGGAUGCAGGUUUCCGACUGGAGAAUCAGGGCGUAGCUGGGUCCGCUGAAGAAGAUAUACCAAAUGGUUAUACCAACUAAGUGGCUUAGGA (((((((((((((....(((((((...((((....))))(((((....))))).....))))))))))))))).((.....))...)))))..(((.....))).(((((...))))).. ( -42.90) >DroGri_CAF1 237 104 - 1 UGGUGGGCCCCAUGAGUUGGCGCCACUUUUGGAUGCAUGACUCCGACUGCAGUAUGAGGGCGUAACAGGGUCCGCUGCAAGCAA--AUCA-AAAUGCAUUAGAU-------------GAA .(((((((((.....((((.((((.(((.....((((..........))))....))))))))))).)))))))))....(((.--....-...))).......-------------... ( -32.10) >DroSec_CAF1 231 120 - 1 UGGUGGGACCCAGAAGACUGCGCCACUUCUGUAUGCAGGCUUCCGAUUGGAGAAUAAGGGCGUAGCUGGGUCCGCUGAUGGAGAUAUACAAAAUGGUUAUACAAUCUAAGUGGCCUCAGA .....((((((((....(((((((.((((((....)))..((((....))))...)))))))))))))))))).((((((........))....((((((.........)))))))))). ( -40.60) >DroSim_CAF1 255 111 - 1 UGGUGGGACCCAGAAGACUGCGCCACUUCUGUAUGCAGGCUUCCGAUUGGAGAAUAAGGGCGUAGCUGGGUCCGCUGAAGGAGAUAUAACAAAUGGUUAUACAAUC---------UCAGA .(((((.((((((....(((((((.((((((....)))..((((....))))...)))))))))))))))))))))....((((((((((.....))))))...))---------))... ( -41.50) >DroEre_CAF1 507 113 - 1 UGGUGGGACCCAGCAGACCACGCCACUUCUGGAUGCAGGCUUCCGACUGGAGAAUCAGCGCGUAGCAGGGUCCGCUGAAAGGGAUA-ACCAGCUUGUUAUGCCAUCUUAAUGCU------ .(((((.((((.(((..(((.........))).)))..(((.(((.(((......))))).).))).)))))))))....((.(((-((......))))).))...........------ ( -33.90) >DroYak_CAF1 255 120 - 1 UGGUGGGACCCAGCAGACUGCGCCACUUUUGGAUGCAGGCUUCCGACUGCAGAAUCAGGGCAUAGCAGGGUCCGCUGAAAGAUAUAUACCAACUUGUUAUAAAAUCUAUAUGCCUGCGAA .(((.((((((.((.....))(((.((..(...(((((........)))))..)..)))))......))))))......((((.((((.(.....).))))..))))....)))...... ( -32.40) >consensus UGGUGGGACCCAGAAGACUGCGCCACUUCUGGAUGCAGGCUUCCGACUGGAGAAUCAGGGCGUAGCAGGGUCCGCUGAAAGAGAUAUACCAAAUGGUUAUACAAUCUAA_UG_C_UCGGA .((((((.(((....(.(((((((...((((....)))).((((....))))......)))))))).)))))))))............................................ (-22.78 = -23.28 + 0.50)

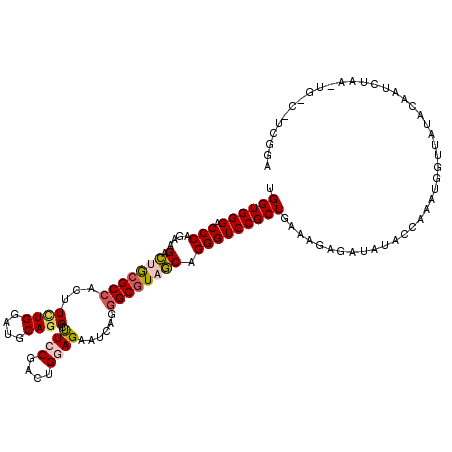

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:22 2006