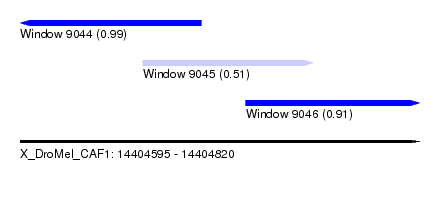
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,404,595 – 14,404,820 |
| Length | 225 |
| Max. P | 0.987005 |

| Location | 14,404,595 – 14,404,697 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.12 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -15.90 |
| Energy contribution | -16.21 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.987005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14404595 102 - 22224390 AUUUCAUUUGCAUGCCGCGCGCAAAGGCAGCGGCGCUGAAAGAAGAAGAACAAGCGGACGGUGUGGA-GCG-GAGCGGUAGCGGCAAAAAGAAAUUAAACACAA (((((.(((((.((((((.(((....(((.((.((((...............))))..)).)))...-)))-..))))))...)))))..)))))......... ( -31.56) >DroSec_CAF1 114602 102 - 1 AUUUCAUUUGCAUGCCGCGCGCAAAGGCAGCGCCGCUGGAAGAAGAAGCAGAAGCGGACGGUGUGGA-GCG-GAGCGGUAGCGGCAAAAAGAAAUUAAACACAA (((((.(((((.((((((.(((....(((.(((((((...............))))).)).)))...-)))-..))))))...)))))..)))))......... ( -35.26) >DroYak_CAF1 114236 82 - 1 AUUUCAUUUGCAUGCCGCGUGCAAAGGCAGCGGCGGUAGAAGAAGA------AGCG----------------GAGCGGUAGCGGCAAAAAGAAAUUAAACACAA .((((.(((((.((((((.(((....)))))))))))))).)))).------.((.----------------..((....)).))................... ( -24.30) >DroAna_CAF1 180746 98 - 1 AUUUCAUUUGCAUGCCGCGCGCAAAGUCAGCAGAGGCAGAGGCAGA------GGCAGAGACUGUGGCAGCAACAGUAGUGGCAGCAAAAAGAAAUUAAACACAA (((((.(((((.((((((((.((.((((.((....((....))...------.))...)))).)))).((....)).)))))))))))..)))))......... ( -28.00) >consensus AUUUCAUUUGCAUGCCGCGCGCAAAGGCAGCGGCGCUAGAAGAAGA______AGCGGACGGUGUGGA_GCG_GAGCGGUAGCGGCAAAAAGAAAUUAAACACAA (((((.(((((.((((((((((....)).))..((((...............))))..................))))))...)))))..)))))......... (-15.90 = -16.21 + 0.31)



| Location | 14,404,664 – 14,404,760 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 85.07 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -27.29 |
| Energy contribution | -27.48 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14404664 96 + 22224390 CCGCUGCCUUUGCGCGCGGCAUGCAAAUGAAAUGUCAUAAAAAUUGCAACUCGGCAGC----------------GGCA-CGGGCAGCGACGUCAAUGUUGCAGCUGCCACUAU ((((((((..(((.....)))(((((((((....)))).....)))))....))))))----------------))..-..((((((..((.......))..))))))..... ( -36.50) >DroSec_CAF1 114671 96 + 1 GCGCUGCCUUUGCGCGCGGCAUGCAAAUGAAAUGUCAUAAAAAUUGCAACUCGGCAGC----------------GGCA-CGGGCAGCGACGUCAAUGUUGCAGCUGCCACUAU .((((((((.(((.((((((((.........)))))........(((......)))))----------------))))-.))))))))........((.((....)).))... ( -34.60) >DroEre_CAF1 99932 96 + 1 CCGCCGCCUUUGCGCGCGGCAUGCAAAUGAAAUGUCAUAAAAAUUGCAACUCGGCAGC----------------GGCA-AGGGCAGCGACGUCAAUGUUGCAGCUGCCACUGU (((((((....))).))))..(((((((((....)))).....)))))....((((((----------------.((.-...)).((((((....)))))).))))))..... ( -35.20) >DroYak_CAF1 114285 102 + 1 CCGCUGCCUUUGCACGCGGCAUGCAAAUGAAAUGUCAUAAAAAUUGCAACUCGGCAGCGGCA----------GCGGCA-CGGGCAGCGACGUCAAUGUUGCAGCUGCCACUGU ((((((((.((((.((.(...(((((((((....)))).....))))).).)))))).))))----------))))..-..((((((..((.......))..))))))..... ( -41.80) >DroAna_CAF1 180811 103 + 1 CUGCUGACUUUGCGCGCGGCAUGCAAAUGAAAUGUCAUAAAAAUUGCAGCUCGGCAGGGAGG----------GGGGCAGCGGGCAGCGGCAUCAAUGUUGCAGCUGCCACUAU (((((..((((.(((.((((.(((((((((....)))).....))))))).)))).).))))----------..)))))..((((((.(((.......))).))))))..... ( -40.80) >DroPer_CAF1 125997 109 + 1 CUGCUGCCUUUGCGCGUGGCAUGCAAAUGAAAUGUCAUAAAAAUUGCAACUCUGCUGCGACGCUGCCGUGCUGCAACG-CUGGCAGCGGCGUCAAUGUUGCAGCUGCCGC--- ..((.((....))))((((((.((..((((....))))......((((((..((..(((.((((((((.((......)-))))))))).)))))..))))))))))))))--- ( -44.40) >consensus CCGCUGCCUUUGCGCGCGGCAUGCAAAUGAAAUGUCAUAAAAAUUGCAACUCGGCAGC________________GGCA_CGGGCAGCGACGUCAAUGUUGCAGCUGCCACUAU ..(((((........))))).(((((((((....)))).....)))))....((((((.................((.....)).((((((....)))))).))))))..... (-27.29 = -27.48 + 0.20)

| Location | 14,404,722 – 14,404,820 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 72.23 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -16.61 |
| Energy contribution | -18.18 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14404722 98 + 22224390 -GGCACGGGCAGCGACGUCAAUGUUGCAGCUGCCACUAUCGCAGCACUGAGAGAAAAGUGC-CAAGCAAA--GGGAUUUUAAGUCA-GCUAUGCAAGUGUGAU- -.((((.((((((((((....)))))).(((((.......))))).............)))-)..(((.(--(.(((.....))).-.)).)))..))))...- ( -31.10) >DroSec_CAF1 114729 103 + 1 -GGCACGGGCAGCGACGUCAAUGUUGCAGCUGCCACUAUCGCAGCACUGAGAGAAAAGUGCACAAGUAAAAAGUAAUUAGAAAUCGAGCGCUAAAAACUAAAUU -(((...((((((..((.......))..))))))......((.(((((........))))).........((....)).........)))))............ ( -24.70) >DroEre_CAF1 99990 85 + 1 -GGCAAGGGCAGCGACGUCAAUGUUGCAGCUGCCACUGUCGCAGCACUGAGAGAAAAGCAG---CGCAAG--U---UGCAAGUUC--ACGCA---AGUU----- -((((..((((((..((.......))..))))))..))))............(((..((((---(....)--)---)))...)))--((...---.)).----- ( -31.40) >DroYak_CAF1 114348 97 + 1 CGGCACGGGCAGCGACGUCAAUGUUGCAGCUGCCACUGUCGCUGCACUAAGAAAAAAGCAC---AGCAAA--AGCAUCUAAAGCU--ACGCAGAAAGUUAAAUU (((((..((((((..((.......))..))))))..)))))((((............((..---.))...--(((.......)))--..))))........... ( -26.00) >consensus _GGCACGGGCAGCGACGUCAAUGUUGCAGCUGCCACUAUCGCAGCACUGAGAGAAAAGCAC___AGCAAA__GG_AUUUAAAGUC__ACGCAGAAAGUUAAAU_ .((((..((((((..((.......))..))))))..))))((.(((((........)))))....))..................................... (-16.61 = -18.18 + 1.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:15 2006