
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,384,289 – 14,384,379 |
| Length | 90 |
| Max. P | 0.941792 |

| Location | 14,384,289 – 14,384,379 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.40 |
| Mean single sequence MFE | -24.06 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.57 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
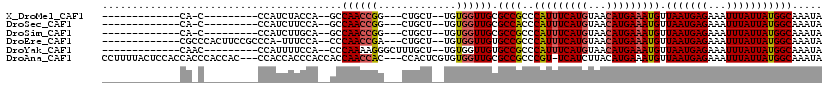
>X_DroMel_CAF1 14384289 90 + 22224390 -------------CA-C---------CCAUCUACCA--GCCAACCGG---CUGCU--UGUGGUUGCGCCGCCCAUUUCAUGUAACAUGAAAUGUUAAUGAGAAAUUUAUUAUGGCAAAUA -------------((-.---------((((....((--(((....))---)))..--.)))).)).((((..(((((((((...))))))))).(((((((...)))))))))))..... ( -25.60) >DroSec_CAF1 100365 90 + 1 -------------CA-C---------CCAUCUUCCA--GCCAACCGG---CUGCU--UGUGGUUGCGCCACCCAUUUCAUGUAACAUGAAAUGUUAAUGAGAAAUUUAUUAUGGCAAAUA -------------((-.---------((((....((--(((....))---)))..--.)))).)).((((..(((((((((...))))))))).(((((((...)))))))))))..... ( -26.30) >DroSim_CAF1 94452 90 + 1 -------------CA-C---------CCAUCUUGCA--GCCAACCGG---CUGCU--UGUGGUUGCGCCGCCCAUUUCAUGUAACAUGAAAUGUUAAUGAGAAAUUUAUUAUGGCAAAUA -------------((-.---------((((...(((--(((....))---)))).--.)))).)).((((..(((((((((...))))))))).(((((((...)))))))))))..... ( -30.10) >DroEre_CAF1 89367 99 + 1 -------------CGCCCACUUCCGCCCA-UUUCCA--CCCAACCGA---CUGCU--UGUGGUUGUGCCGCCCAUUUCAUGUAACAUGAAAUGUUAAUGAGAAAUUUAUUAUGGCAAAUA -------------................-......--..(((((.(---(....--.)))))))(((((..(((((((((...))))))))).(((((((...)))))))))))).... ( -20.90) >DroYak_CAF1 101969 94 + 1 -------------CAAC---------CCAUUUUCCA--CCCAAAAGGGCUUUGCU--UGUGGUUGUGCCGCCCAUUUCAUGUAACAUGAAAUGUUAAUGAGAAAUUUAUUAUGGCAAAUA -------------((((---------(((.......--(((....))).......--)).)))))(((((..(((((((((...))))))))).(((((((...)))))))))))).... ( -24.94) >DroAna_CAF1 165834 113 + 1 CCUUUUACUCCACCACCCACCAC---CCACCACCCACCACCAACCAC---CCACUCGUGUGGUUGCGCCGCCCGU-UCAUCUUACAUGAAAUGUUAAUGAGAAAUUUAUUAUGGCAAAUA .......................---..............(((((((---.(....).))))))).((((....(-((((.....)))))....(((((((...)))))))))))..... ( -16.50) >consensus _____________CA_C_________CCAUCUUCCA__GCCAACCGG___CUGCU__UGUGGUUGCGCCGCCCAUUUCAUGUAACAUGAAAUGUUAAUGAGAAAUUUAUUAUGGCAAAUA ........................................((((((.............)))))).((((..(((((((((...))))))))).(((((((...)))))))))))..... (-16.32 = -16.57 + 0.25)

| Location | 14,384,289 – 14,384,379 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.40 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -14.15 |
| Energy contribution | -14.76 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14384289 90 - 22224390 UAUUUGCCAUAAUAAAUUUCUCAUUAACAUUUCAUGUUACAUGAAAUGGGCGGCGCAACCACA--AGCAG---CCGGUUGGC--UGGUAGAUGG---------G-UG------------- (((((((((..................(((((((((...)))))))))..((((((.......--.)).)---)))......--))))))))).---------.-..------------- ( -26.10) >DroSec_CAF1 100365 90 - 1 UAUUUGCCAUAAUAAAUUUCUCAUUAACAUUUCAUGUUACAUGAAAUGGGUGGCGCAACCACA--AGCAG---CCGGUUGGC--UGGAAGAUGG---------G-UG------------- ...................((((((..(((((((((...))))))))).((((.....)))).--..(((---((....)))--))...)))))---------)-..------------- ( -25.80) >DroSim_CAF1 94452 90 - 1 UAUUUGCCAUAAUAAAUUUCUCAUUAACAUUUCAUGUUACAUGAAAUGGGCGGCGCAACCACA--AGCAG---CCGGUUGGC--UGCAAGAUGG---------G-UG------------- ...................((((((..(((((((((...))))))))).(((((.(((((...--.....---..)))))))--)))..)))))---------)-..------------- ( -28.50) >DroEre_CAF1 89367 99 - 1 UAUUUGCCAUAAUAAAUUUCUCAUUAACAUUUCAUGUUACAUGAAAUGGGCGGCACAACCACA--AGCAG---UCGGUUGGG--UGGAAA-UGGGCGGAAGUGGGCG------------- .....(((.......(((((.((((..(((((((((...))))))))).......(((((((.--....)---).)))))))--))))))-)..((....)).))).------------- ( -29.00) >DroYak_CAF1 101969 94 - 1 UAUUUGCCAUAAUAAAUUUCUCAUUAACAUUUCAUGUUACAUGAAAUGGGCGGCACAACCACA--AGCAAAGCCCUUUUGGG--UGGAAAAUGG---------GUUG------------- ....((((.((((.........)))).(((((((((...)))))))))...))))(((((.((--......((((....)))--)......)))---------))))------------- ( -26.10) >DroAna_CAF1 165834 113 - 1 UAUUUGCCAUAAUAAAUUUCUCAUUAACAUUUCAUGUAAGAUGA-ACGGGCGGCGCAACCACACGAGUGG---GUGGUUGGUGGUGGGUGGUGG---GUGGUGGGUGGUGGAGUAAAAGG ..((..((((.....((..(((((((.(..(((((.....))))-)...((.((.(((((((........---))))))))).)).).))))))---)..))..))))..))........ ( -32.10) >consensus UAUUUGCCAUAAUAAAUUUCUCAUUAACAUUUCAUGUUACAUGAAAUGGGCGGCGCAACCACA__AGCAG___CCGGUUGGC__UGGAAGAUGG_________G_UG_____________ ....((((.((((.........)))).(((((((((...)))))))))...))))(((((...............)))))........................................ (-14.15 = -14.76 + 0.61)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:05 2006