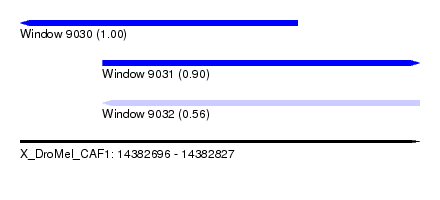
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,382,696 – 14,382,827 |
| Length | 131 |
| Max. P | 0.998486 |

| Location | 14,382,696 – 14,382,787 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 64.46 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -12.73 |
| Energy contribution | -15.60 |
| Covariance contribution | 2.87 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.89 |
| Structure conservation index | 0.41 |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.998486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14382696 91 - 22224390 UUCUUUGGCUGGCCAUAAAAUUCUGCUCAAUUUGCAUGGCCAACUUGGC--CAGCGAA-----CUGAAGUGGCCAAGUAAACAAAAACUUGGCUAACG (((.((.((((((((......(((((.......))).))......))))--)))).))-----..))).(((((((((........)))))))))... ( -33.60) >DroYak_CAF1 100606 96 - 1 UCAUUUAGUAGGCCAUCACAUUCUGCUCAAUUUGCGUGGCUAACUUGGC--CAGCGAAGGCCACUGAAGUGGCCAAGUAAACAAAAACUUGGCCAACG ...((((((.((((.((.(.....((.......)).(((((.....)))--))).)).)))))))))).(((((((((........)))))))))... ( -37.70) >DroAna_CAF1 164671 83 - 1 UCGUUUACUCGGCCCUCAAAU---------UUUGUGUUGCCUACUUGGCAGCGCCGAC-----UUCGAGUAUCUAAGCGAAAACAAAACUUUCUAAG- (((((((...((..(((.((.---------((.((((((((.....)))))))).)).-----)).)))..))))))))).................- ( -22.60) >consensus UCAUUUAGUAGGCCAUCAAAUUCUGCUCAAUUUGCGUGGCCAACUUGGC__CAGCGAA_____CUGAAGUGGCCAAGUAAACAAAAACUUGGCUAACG ..........((((((((((((......)))))).))))))............................(((((((((........)))))))))... (-12.73 = -15.60 + 2.87)


| Location | 14,382,723 – 14,382,827 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 73.38 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -10.69 |
| Energy contribution | -12.75 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14382723 104 + 22224390 CACUUCAG-----UUCGCUGGCCAAGUUGGCCAUGCAAAUUGAGCAGAAUUUUAUGGCCAGCCAAAGAAGGCUAAUUUGGUAAAUUUUUUUUUACUUGGCUUAAAAUUU ...(((((-----((.(((((((.....))))).)).)))))))..((((((((.((((((((......)))).....((((((......)))))).)))))))))))) ( -35.10) >DroSec_CAF1 98834 81 + 1 CACUUCAG-----UUCGCUGG-----------------------CAGAGUUUUAUGGCCAGCUAAAGAAGGCUAAUUUCGUAAAUUUAUUUUUACUUGGUUUAAAAUUU ..((((..-----((.(((((-----------------------(...........)))))).)).))))(((((....(((((......))))))))))......... ( -17.10) >DroSim_CAF1 92933 81 + 1 CACUUCAG-----UUCGCUGG-----------------------CAGAAUUUUAUGGCCAGCUAAAGAAGGCUAAUUUCGUAAAUUUAUUUUUACUUGGCUUAAAAUUU ..((((..-----((.(((((-----------------------(...........)))))).)).))))(((((....(((((......))))))))))......... ( -19.20) >DroYak_CAF1 100633 107 + 1 CACUUCAGUGGCCUUCGCUGGCCAAGUUAGCCACGCAAAUUGAGCAGAAUGUGAUGGCCUACUAAAUGAGGCUAAUUUUGUAAAUUUGUU--CACUUGGCUUACAAAUG ........(((((......))))).((.(((((.(((((((..(((((((....((((((........))))))))))))).))))))).--....))))).))..... ( -33.70) >consensus CACUUCAG_____UUCGCUGG_______________________CAGAAUUUUAUGGCCAGCUAAAGAAGGCUAAUUUCGUAAAUUUAUUUUUACUUGGCUUAAAAUUU ................(((((((................................)))))))......(((((((....(((((......))))))))))))....... (-10.69 = -12.75 + 2.06)


| Location | 14,382,723 – 14,382,827 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 73.38 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -8.69 |
| Energy contribution | -10.56 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14382723 104 - 22224390 AAAUUUUAAGCCAAGUAAAAAAAAAUUUACCAAAUUAGCCUUCUUUGGCUGGCCAUAAAAUUCUGCUCAAUUUGCAUGGCCAACUUGGCCAGCGAA-----CUGAAGUG .(((((((.(((..(((((......)))))......((((......)))))))..)))))))..(((((.(((((.(((((.....))))))))))-----.)).))). ( -31.40) >DroSec_CAF1 98834 81 - 1 AAAUUUUAAACCAAGUAAAAAUAAAUUUACGAAAUUAGCCUUCUUUAGCUGGCCAUAAAACUCUG-----------------------CCAGCGAA-----CUGAAGUG ..............(((((......))))).........((((....((((((...........)-----------------------)))))...-----..)))).. ( -15.80) >DroSim_CAF1 92933 81 - 1 AAAUUUUAAGCCAAGUAAAAAUAAAUUUACGAAAUUAGCCUUCUUUAGCUGGCCAUAAAAUUCUG-----------------------CCAGCGAA-----CUGAAGUG ..............(((((......))))).........((((....((((((...........)-----------------------)))))...-----..)))).. ( -15.80) >DroYak_CAF1 100633 107 - 1 CAUUUGUAAGCCAAGUG--AACAAAUUUACAAAAUUAGCCUCAUUUAGUAGGCCAUCACAUUCUGCUCAAUUUGCGUGGCUAACUUGGCCAGCGAAGGCCACUGAAGUG ..((((((((....(..--..)...))))))))..........((((((.((((.((.(.....((.......)).(((((.....)))))).)).))))))))))... ( -26.80) >consensus AAAUUUUAAGCCAAGUAAAAAUAAAUUUACGAAAUUAGCCUUCUUUAGCUGGCCAUAAAAUUCUG_______________________CCAGCGAA_____CUGAAGUG ..............(((((......)))))....((((.....(((.((((((((..............................))))))))))).....)))).... ( -8.69 = -10.56 + 1.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:03 2006