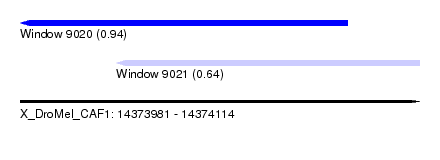
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,373,981 – 14,374,114 |
| Length | 133 |
| Max. P | 0.935489 |

| Location | 14,373,981 – 14,374,090 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.47 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -25.42 |
| Energy contribution | -25.50 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14373981 109 - 22224390 CUGUAUCCAGAAUCCUGUA--UUCUGUAUCCCGAAUCCUGGGUUGACAGGGCAGGGAUUACGCUUAAUAUGGUGUGAACAUUUAUUUAAAUGCUUUAAAAGCACUGCCAUU ((((...((((((.....)--)))))...((((.....))))...))))(((((...(((((((......))))))).............((((.....)))))))))... ( -30.10) >DroSec_CAF1 90144 109 - 1 CUGUAUCCAGAAUCCUGCA--UUCUGUAUCCCGAAUCCCGGGUUGACAGGGCAGGGAUUAUGCUUAAUAUGGUGUGAACAUUUAUUUAAAUGCUUUAAAAGCACUACCAUU (((....)))..((((((.--..((((..((((.....))))...)))).)))))).(((((((......))))))).............((((.....))))........ ( -28.80) >DroSim_CAF1 84283 109 - 1 CUGUAUCCAGAAUCCUGCA--UUCUGUAUCCCGAAUCCCGGGUUGACAGGGCAGGGAUUAUGCUUAAUAUGGUGUGAACAUUUAUUUAAAUGCUUUAAAAGCACUGCCAUU ((((...((((((.....)--)))))...((((.....))))...))))(((((...(((((((......))))))).............((((.....)))))))))... ( -29.70) >DroEre_CAF1 81288 99 - 1 ---------GAAUCCCGCA--UUCUGCAUCCC-AAUCCUCGGCUGACAGGGCGGGGAUUACGCCUAAUAUGGUGUGAACAUUUAUUCAAAUGCUGCAGAAGCACUGCCACU ---------.......((.--(((((((....-..((((((.((....)).))))))(((((((......))))))).(((((....))))).)))))))))......... ( -32.90) >DroYak_CAF1 91650 92 - 1 -------------------AGUUCUGUAUCCCGAAUCCUGGGCUGACAGGGCAGGGAUUACGCUUAAUGCGGUGUGAACAUUUAUUUAAAUGCUUUAAAAGCACUGCCACU -------------------....((((..((((.....))))...))))(((((...(((((((......))))))).............((((.....)))))))))... ( -27.00) >consensus CUGUAUCCAGAAUCCUGCA__UUCUGUAUCCCGAAUCCUGGGUUGACAGGGCAGGGAUUACGCUUAAUAUGGUGUGAACAUUUAUUUAAAUGCUUUAAAAGCACUGCCAUU .......................((((..((((.....))))...))))(((((...(((((((......))))))).............((((.....)))))))))... (-25.42 = -25.50 + 0.08)



| Location | 14,374,013 – 14,374,114 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -24.35 |
| Energy contribution | -23.98 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14374013 101 - 22224390 UGACCA--AAGU------UCUGGACCGAAGUUCUGUAUCCAGAAUCCUGUAUUCUGUAUCCCGAAUCCUGGGUUGACAGGGCAGGGAUUACGCUUAAUAUGGUGUGAAC ......--..((------((((((.............))))))))(((((..(((((..((((.....))))...))))))))))..(((((((......))))))).. ( -31.52) >DroSec_CAF1 90176 101 - 1 UGACCA--AAGU------CCAGGACCGAAGUUCUGUAUCCAGAAUCCUGCAUUCUGUAUCCCGAAUCCCGGGUUGACAGGGCAGGGAUUAUGCUUAAUAUGGUGUGAAC .(((..--..))------)((((((....)))))).........((((((...((((..((((.....))))...)))).)))))).(((((((......))))))).. ( -31.80) >DroSim_CAF1 84315 101 - 1 UGACCA--AAGU------CCUGGACCGAAGUUCUGUAUCCAGAAUCCUGCAUUCUGUAUCCCGAAUCCCGGGUUGACAGGGCAGGGAUUAUGCUUAAUAUGGUGUGAAC ...(((--....------..)))......((((((....))))))(((((...((((..((((.....))))...)))).)))))..(((((((......))))))).. ( -31.10) >DroEre_CAF1 81320 95 - 1 UGGCUAGGAGCUCUGCAUCCAGAAUCCA-------------GAAUCCCGCAUUCUGCAUCCC-AAUCCUCGGCUGACAGGGCGGGGAUUACGCCUAAUAUGGUGUGAAC ......(((..((((....)))).))).-------------...((((((...(((((.((.-.......)).)).))).)))))).(((((((......))))))).. ( -32.40) >consensus UGACCA__AAGU______CCAGGACCGAAGUUCUGUAUCCAGAAUCCUGCAUUCUGUAUCCCGAAUCCCGGGUUGACAGGGCAGGGAUUACGCUUAAUAUGGUGUGAAC ............................................((((((...((((..((((.....))))...)))).)))))).(((((((......))))))).. (-24.35 = -23.98 + -0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:53 2006