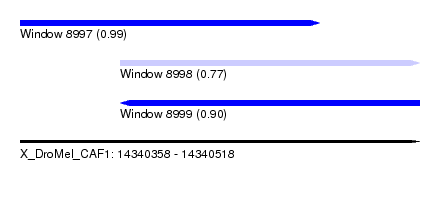
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,340,358 – 14,340,518 |
| Length | 160 |
| Max. P | 0.992513 |

| Location | 14,340,358 – 14,340,478 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.38 |
| Mean single sequence MFE | -41.98 |
| Consensus MFE | -35.35 |
| Energy contribution | -35.35 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
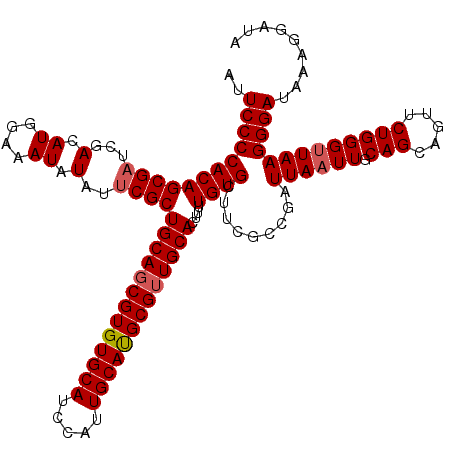
>X_DroMel_CAF1 14340358 120 + 22224390 AUUCAGAAUUCAGCGGAAGGGGGGCCGAGCAUAAAUUGCAAUUCCCCACAGCGAUCGACAUGGAAAUAUAUUCGCUGCAGCGUGUGCAUGCAAUGCAUGCGUUGCACUUUUGUGCUCCGC ............(((((.((((((....(((.....)))..)))))).((((((...(.((....)).)..)))))).(((((((((((...)))))))))))((((....))))))))) ( -43.30) >DroSec_CAF1 52369 119 + 1 AUUCAGAAUUCAGCGGAAGGGGG-CCGAGCAUAAAUUGCAAUUCCCCACAGCGAUCGACAUGGAAAUAUAUUCGCUGCAGCGUGUGCAUCCAUUGCAUGCGUUGCACUUUUGUGCUUCGC .....(((....(((((((((((-....(((.....)))....))))..(((((...(.((....)).)..)))))((((((((((((.....)))))))))))).)))))))..))).. ( -42.70) >DroSim_CAF1 50078 118 + 1 AUUCAGAAUUCAGCGGAAGGGGG--CGAGCAUAAAUUGCAAUUCCCCACAGCGAUCGACAUGGAAAUAUAUUCGCUGCAGCGUGUGCAUCCAUUGCAUGCGUUGCACUUUUGUGCUUCGC .....(((....(((((((((((--...(((.....)))....))))..(((((...(.((....)).)..)))))((((((((((((.....)))))))))))).)))))))..))).. ( -42.30) >DroEre_CAF1 51308 118 + 1 AUUCAGAACUCGGCGGAAG-GGG-CCGAGCAUAAAUUGCCAUUCCCCACAGCGAUCGACAUGGAAAUAUAUGCGCUGCAGCGUGUGCAUCCACUGCACGCGUUGCAGUUUUGUGCCUCGC ..................(-(((-(((((((((.(((.((((((............)).)))).))).)))))(((((((((((((((.....))))))))))))))).))).))))).. ( -45.50) >DroYak_CAF1 54911 119 + 1 AUUCAGAAUUCAGCGGAUGAGGG-CAGAGCAUAAAUUGCAGUUCCCCACAGCGAUCGACAUGGAAAUAUAUUCGCUGCAUCGUGUGCAUCCAUUGCAUGCGUUGCACUUUUGUGCGCCGC ............((((.((.(((-....(((.....)))....)))))((((((...(.((....)).)..))))))...((((((((.....))))))))..((((....)))).)))) ( -36.10) >consensus AUUCAGAAUUCAGCGGAAGGGGG_CCGAGCAUAAAUUGCAAUUCCCCACAGCGAUCGACAUGGAAAUAUAUUCGCUGCAGCGUGUGCAUCCAUUGCAUGCGUUGCACUUUUGUGCUUCGC ............((((....(((.....(((.....)))....)))((((((((.................))))(((((((((((((.....)))))))))))))....))))..)))) (-35.35 = -35.35 + 0.00)


| Location | 14,340,398 – 14,340,518 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -38.28 |
| Consensus MFE | -33.23 |
| Energy contribution | -33.67 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.765953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14340398 120 + 22224390 AUUCCCCACAGCGAUCGACAUGGAAAUAUAUUCGCUGCAGCGUGUGCAUGCAAUGCAUGCGUUGCACUUUUGUGCUCCGCCGAUUAAUUGCAGCAGUUCUGGGCUAAGGGAUAAAGGAUA ..((((((((((((...(.((....)).)..))))((((((((((((((...))))))))))))))....))))....(((.(..(((((...))))).).)))...))))......... ( -37.20) >DroSec_CAF1 52408 120 + 1 AUUCCCCACAGCGAUCGACAUGGAAAUAUAUUCGCUGCAGCGUGUGCAUCCAUUGCAUGCGUUGCACUUUUGUGCUUCGCCGAUUAAUUGCAGUAGUUCUGGGUUAAGGGAUAAAGGAUA ..((((((((((((...(.((....)).)..))))(((((((((((((.....)))))))))))))....)))).........((((((.(((.....)))))))))))))......... ( -36.40) >DroSim_CAF1 50116 120 + 1 AUUCCCCACAGCGAUCGACAUGGAAAUAUAUUCGCUGCAGCGUGUGCAUCCAUUGCAUGCGUUGCACUUUUGUGCUUCGCCGAUUAAUUGCAGCAGUUCUGGGUUAAGGGAUAAAGGAUA ....((((.(((..........(((.....)))(((((((((((((((.....))))))))))((((....))))..............))))).))).))))................. ( -36.10) >DroEre_CAF1 51346 120 + 1 AUUCCCCACAGCGAUCGACAUGGAAAUAUAUGCGCUGCAGCGUGUGCAUCCACUGCACGCGUUGCAGUUUUGUGCCUCGCCGAUUAAUUGCAGCGGCGCUGGGUUAAGGGAUACAGGAUA ..(((((((((.....(.((((......)))))(((((((((((((((.....))))))))))))))).)))))((.(((((...........)))))..)).....))))......... ( -45.80) >DroYak_CAF1 54950 120 + 1 GUUCCCCACAGCGAUCGACAUGGAAAUAUAUUCGCUGCAUCGUGUGCAUCCAUUGCAUGCGUUGCACUUUUGUGCGCCGCCGAUUAAUUGCAGCGCUUCUGGGUUAAGGGAUAAAGGAUA ....((((.(((..........(((.....)))(((((((((.(((((.....)))))(((.(((((....))))).)))))).....)))))))))..))))................. ( -35.90) >consensus AUUCCCCACAGCGAUCGACAUGGAAAUAUAUUCGCUGCAGCGUGUGCAUCCAUUGCAUGCGUUGCACUUUUGUGCUUCGCCGAUUAAUUGCAGCAGUUCUGGGUUAAGGGAUAAAGGAUA ..((((((((((((.................))))(((((((((((((.....)))))))))))))....)))).........((((((.(((.....)))))))))))))......... (-33.23 = -33.67 + 0.44)


| Location | 14,340,398 – 14,340,518 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -35.98 |
| Consensus MFE | -29.96 |
| Energy contribution | -30.12 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
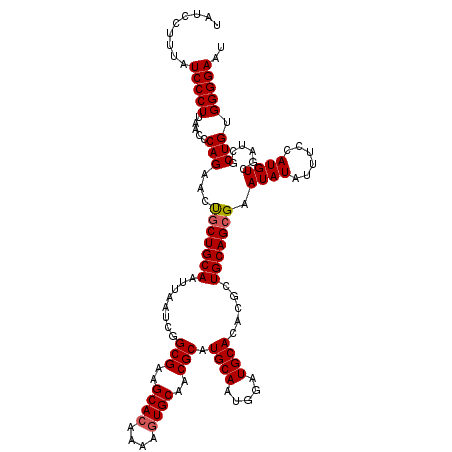
>X_DroMel_CAF1 14340398 120 - 22224390 UAUCCUUUAUCCCUUAGCCCAGAACUGCUGCAAUUAAUCGGCGGAGCACAAAAGUGCAACGCAUGCAUUGCAUGCACACGCUGCAGCGAAUAUAUUUCCAUGUCGAUCGCUGUGGGGAAU .........(((((.(((......((((((........)))))).((((....))))...((((((...))))))....)))(((((((..((((....))))...)))))))))))).. ( -39.30) >DroSec_CAF1 52408 120 - 1 UAUCCUUUAUCCCUUAACCCAGAACUACUGCAAUUAAUCGGCGAAGCACAAAAGUGCAACGCAUGCAAUGGAUGCACACGCUGCAGCGAAUAUAUUUCCAUGUCGAUCGCUGUGGGGAAU .........(((((...............((.........(((..((((....))))..))).((((.....))))...)).(((((((..((((....))))...)))))))))))).. ( -31.70) >DroSim_CAF1 50116 120 - 1 UAUCCUUUAUCCCUUAACCCAGAACUGCUGCAAUUAAUCGGCGAAGCACAAAAGUGCAACGCAUGCAAUGGAUGCACACGCUGCAGCGAAUAUAUUUCCAUGUCGAUCGCUGUGGGGAAU .........(((((.....(((....((((((........(((..((((....))))..))).((((.....)))).....))))))(((.....)))...........))).))))).. ( -32.30) >DroEre_CAF1 51346 120 - 1 UAUCCUGUAUCCCUUAACCCAGCGCCGCUGCAAUUAAUCGGCGAGGCACAAAACUGCAACGCGUGCAGUGGAUGCACACGCUGCAGCGCAUAUAUUUCCAUGUCGAUCGCUGUGGGGAAU .........(((((.....(((((..((((((........(((..(((......)))..)))(((((.....)))))....))))))((((........))))....))))).))))).. ( -39.90) >DroYak_CAF1 54950 120 - 1 UAUCCUUUAUCCCUUAACCCAGAAGCGCUGCAAUUAAUCGGCGGCGCACAAAAGUGCAACGCAUGCAAUGGAUGCACACGAUGCAGCGAAUAUAUUUCCAUGUCGAUCGCUGUGGGGAAC .................((((..(((((((((.....((((((..((((....))))..))).((((.....))))..)))))))))(((.....)))..........))).)))).... ( -36.70) >consensus UAUCCUUUAUCCCUUAACCCAGAACUGCUGCAAUUAAUCGGCGAAGCACAAAAGUGCAACGCAUGCAAUGGAUGCACACGCUGCAGCGAAUAUAUUUCCAUGUCGAUCGCUGUGGGGAAU .........(((((.....(((...(((((((........(((..((((....))))..))).((((.....)))).....))))))).((((......))))......))).))))).. (-29.96 = -30.12 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:33 2006