
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,337,999 – 14,338,129 |
| Length | 130 |
| Max. P | 0.988730 |

| Location | 14,337,999 – 14,338,090 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 95.38 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -23.58 |
| Energy contribution | -24.22 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14337999 91 - 22224390 CAUCAUAACUGCCAAUGCGAAACGCGUGCUGAAGGCAGCCGAAAAUGCGAAAAUUUUGGGCAUGCAUCAGUCAACCGGGCGGAGAAAGAGA ........(((((.(((((...)))))(((((..((((((.(((((......))))).))).))).)))))......)))))......... ( -30.10) >DroSec_CAF1 50009 91 - 1 CAUCAUAACUGCCAAUGCGAAACGCGUGCUGAAGGCAGCCGAAAAUGCGAAAAUUUUGGGCAUGCAUCAGUCAACCGGACGGAGAAAGAGA ........(((((.(((((...)))))(((((..((((((.(((((......))))).))).))).))))).....)).)))......... ( -26.60) >DroSim_CAF1 48033 91 - 1 CAUCAUAACUGCCAAUGCGAAACGCGUGCUGAAGGCAGCCGAAAAUGCGAAAAUUUUGGGCAUGCAUCAGUCAACCGGACGGAGAAAGAGA ........(((((.(((((...)))))(((((..((((((.(((((......))))).))).))).))))).....)).)))......... ( -26.60) >DroEre_CAF1 49013 91 - 1 CAUCAUAACUGCCAAUGCGAAACGCGUGCUGAAGGCAGCCGAAAGUUCGAAAAUGCUGGGCAUGCAUCAGUCAACCGGACGGAGAAAGAGA ........(((((.(((((...)))))(((((..((((((...(((........))).))).))).))))).....)).)))......... ( -23.30) >DroYak_CAF1 52511 90 - 1 CAUCAUAACUGCCGAUGCGAAACGCGUGCUGAAGGCAGCCGAAAAUGCGAAAAUGUUG-GCAUGCAUCAGUCCGCCGGACGGCGAAAGAGA .........(((((((((......((.((((....))))))...((((.((....)).-))))))))).((((...))))))))....... ( -28.30) >consensus CAUCAUAACUGCCAAUGCGAAACGCGUGCUGAAGGCAGCCGAAAAUGCGAAAAUUUUGGGCAUGCAUCAGUCAACCGGACGGAGAAAGAGA ........(((((.(((((...)))))(((((..((((((.(((((......))))).))).))).))))).....)).)))......... (-23.58 = -24.22 + 0.64)

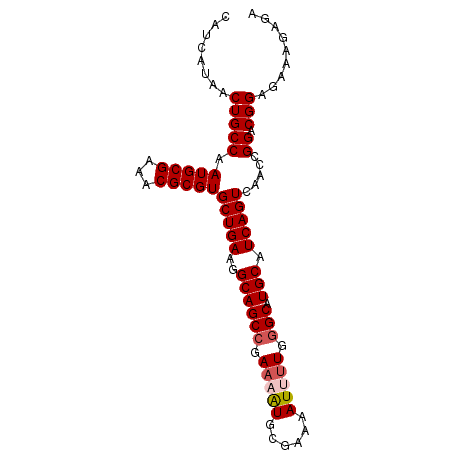
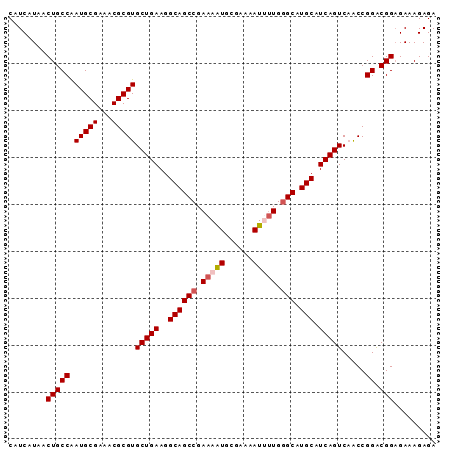
| Location | 14,338,039 – 14,338,129 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 81.44 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -17.69 |
| Energy contribution | -18.44 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14338039 90 + 22224390 UUCGCAUUUUCGGCUGCCUUCAGCACGCGUUUCGCAUUGGCAGUUAUGAUGCGGUUGCCUUGGCUUCCUCCGUUCUCGCAUCAUUUGCAA ...(((.....(((((((....((.........))...)))))))(((((((((..((...((...))...))..))))))))).))).. ( -27.20) >DroSec_CAF1 50049 90 + 1 UUCGCAUUUUCGGCUGCCUUCAGCACGCGUUUCGCAUUGGCAGUUAUGAUGCGGUUGCCUUGGCUUCCUUCGUUCUCGCAUCAUUUGCAA ...(((.....(((((((....((.........))...)))))))(((((((((..((...((...))...))..))))))))).))).. ( -27.20) >DroSim_CAF1 48073 90 + 1 UUCGCAUUUUCGGCUGCCUUCAGCACGCGUUUCGCAUUGGCAGUUAUGAUGCNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN ...(((((...(((((((....((.........))...)))))))..)))))...................................... ( -15.60) >DroEre_CAF1 49053 90 + 1 UUCGAACUUUCGGCUGCCUUCAGCACGCGUUUCGCAUUGGCAGUUAUGAUGCGGUUGCCUUGGCUUCCUUCGUUCUCGCAUCAUUUGCAA ...((((....((..(((..((((.((((((.(((....)).)....))))))))))....)))..))...))))..(((.....))).. ( -24.50) >DroYak_CAF1 52550 90 + 1 UUCGCAUUUUCGGCUGCCUUCAGCACGCGUUUCGCAUCGGCAGUUAUGAUGCGGUUGCCUUGGCUUCCUUCGUUCUCGCAUCAUUUGCAA ...(((.....(((((((....((.........))...)))))))(((((((((..((...((...))...))..))))))))).))).. ( -26.50) >consensus UUCGCAUUUUCGGCUGCCUUCAGCACGCGUUUCGCAUUGGCAGUUAUGAUGCGGUUGCCUUGGCUUCCUUCGUUCUCGCAUCAUUUGCAA ...(((((...(((((((....((.........))...)))))))..)))))((..((....))..)).........(((.....))).. (-17.69 = -18.44 + 0.75)



| Location | 14,338,039 – 14,338,129 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 81.44 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -16.58 |
| Energy contribution | -19.38 |
| Covariance contribution | 2.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14338039 90 - 22224390 UUGCAAAUGAUGCGAGAACGGAGGAAGCCAAGGCAACCGCAUCAUAACUGCCAAUGCGAAACGCGUGCUGAAGGCAGCCGAAAAUGCGAA (((((.((((((((.....((......))..(....)))))))))..(((((.(((((...)))))......))))).......))))). ( -28.30) >DroSec_CAF1 50049 90 - 1 UUGCAAAUGAUGCGAGAACGAAGGAAGCCAAGGCAACCGCAUCAUAACUGCCAAUGCGAAACGCGUGCUGAAGGCAGCCGAAAAUGCGAA (((((.((((((((....(....)..((....))...))))))))..(((((.(((((...)))))......))))).......))))). ( -29.50) >DroSim_CAF1 48073 90 - 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGCAUCAUAACUGCCAAUGCGAAACGCGUGCUGAAGGCAGCCGAAAAUGCGAA ......................................((((.....(((((.(((((...)))))......)))))......))))... ( -14.00) >DroEre_CAF1 49053 90 - 1 UUGCAAAUGAUGCGAGAACGAAGGAAGCCAAGGCAACCGCAUCAUAACUGCCAAUGCGAAACGCGUGCUGAAGGCAGCCGAAAGUUCGAA ..(((.((((((((....(....)..((....))...))))))))...))).....((((...((.((((....))))))....)))).. ( -27.30) >DroYak_CAF1 52550 90 - 1 UUGCAAAUGAUGCGAGAACGAAGGAAGCCAAGGCAACCGCAUCAUAACUGCCGAUGCGAAACGCGUGCUGAAGGCAGCCGAAAAUGCGAA (((((.((((((((....(....)..((....))...))))))))..(((((.(((((...)))))......))))).......))))). ( -29.50) >consensus UUGCAAAUGAUGCGAGAACGAAGGAAGCCAAGGCAACCGCAUCAUAACUGCCAAUGCGAAACGCGUGCUGAAGGCAGCCGAAAAUGCGAA (((((.((((((((....(....)..((....))...))))))))..(((((.(((((...)))))......))))).......))))). (-16.58 = -19.38 + 2.80)

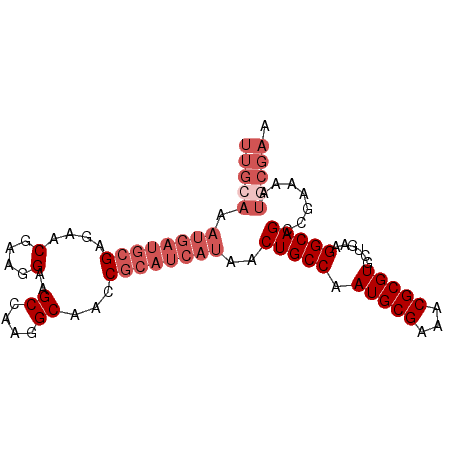
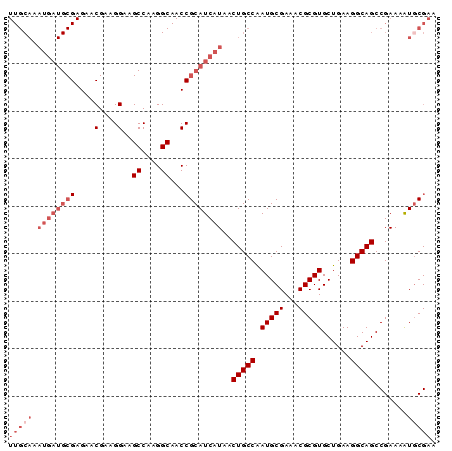
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:31 2006