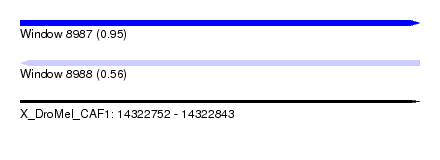
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,322,752 – 14,322,843 |
| Length | 91 |
| Max. P | 0.951627 |

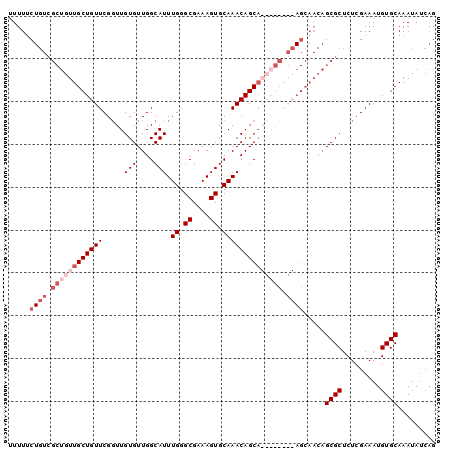
| Location | 14,322,752 – 14,322,843 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.97 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -22.69 |
| Energy contribution | -26.00 |
| Covariance contribution | 3.31 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14322752 91 + 22224390 GUUUUCUGUCGCUGUUGCUGUUCGGUUGUGUUGGCAUUUGGGCGAAAGUGCAAACAGC---------AGCAACAGCGCUCUCGAAAUGUGCAAAUAUCAG (((((..(.(((((((((((((.((((.....))).((((.((....)).))))))))---------))))))))))..)..)))))............. ( -30.70) >DroSec_CAF1 35206 100 + 1 UUUUUCUGUCGCUGUUGCUGUUCGGUUGUGUUGGCAUUUGGGCGAAAGUGCAAACAGCAGCAGCAGCAGCAACAGCGCUCUCGAAAUGUGCAAAUAUCAG ......(((.(((((((((((...(((((....((((((......))))))..))))).))))))))))).)))((((.........))))......... ( -39.30) >DroSim_CAF1 32580 100 + 1 UUUUUCUGUCGCUGUUGCUGUUCGGUUGUGUUGGCAUUUGGGCGAAAGUGCAAACAGCAGCAGCAGCAGCAACAGCGCUCUCGAAAUGUGCAAAUAUCAG ......(((.(((((((((((...(((((....((((((......))))))..))))).))))))))))).)))((((.........))))......... ( -39.30) >DroEre_CAF1 33516 79 + 1 UUUUGC------CGUUGCUGUUCGCUUGUGUUGGCAUUUGGGCGAAAGUGCAAACAGCA---------------GCGCUCUCGAAAUGUGCAAAUAGCAG ...(((------..((((..((((...((((((...((((.((....)).))))...))---------------))))...))))....))))...))). ( -26.40) >consensus UUUUUCUGUCGCUGUUGCUGUUCGGUUGUGUUGGCAUUUGGGCGAAAGUGCAAACAGCA________AGCAACAGCGCUCUCGAAAUGUGCAAAUAUCAG .....((((.((((((((((((.....(((....))).((.((....)).)))))))))))))).)))).....((((.........))))......... (-22.69 = -26.00 + 3.31)


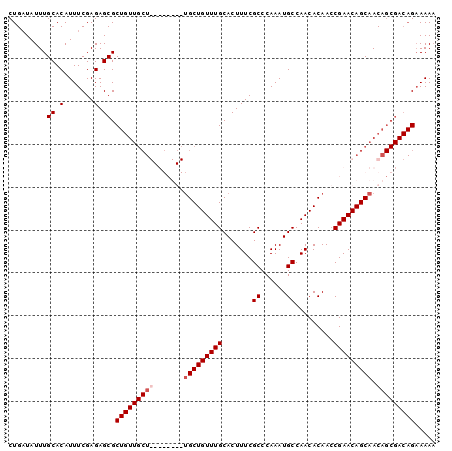
| Location | 14,322,752 – 14,322,843 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 84.97 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -15.49 |
| Energy contribution | -16.87 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14322752 91 - 22224390 CUGAUAUUUGCACAUUUCGAGAGCGCUGUUGCU---------GCUGUUUGCACUUUCGCCCAAAUGCCAACACAACCGAACAGCAACAGCGACAGAAAAC ..............((((.....((((((((((---------(.((((.(((.((......)).))).))))........)))))))))))...)))).. ( -23.90) >DroSec_CAF1 35206 100 - 1 CUGAUAUUUGCACAUUUCGAGAGCGCUGUUGCUGCUGCUGCUGCUGUUUGCACUUUCGCCCAAAUGCCAACACAACCGAACAGCAACAGCGACAGAAAAA ..............((((.....(((((((((((...(.(.((.((((.(((.((......)).))).)))))).).)..)))))))))))...)))).. ( -27.80) >DroSim_CAF1 32580 100 - 1 CUGAUAUUUGCACAUUUCGAGAGCGCUGUUGCUGCUGCUGCUGCUGUUUGCACUUUCGCCCAAAUGCCAACACAACCGAACAGCAACAGCGACAGAAAAA ..............((((.....(((((((((((...(.(.((.((((.(((.((......)).))).)))))).).)..)))))))))))...)))).. ( -27.80) >DroEre_CAF1 33516 79 - 1 CUGCUAUUUGCACAUUUCGAGAGCGC---------------UGCUGUUUGCACUUUCGCCCAAAUGCCAACACAAGCGAACAGCAACG------GCAAAA ......(((((......(....)((.---------------((((((((((......((......))........)))))))))).))------))))). ( -20.74) >consensus CUGAUAUUUGCACAUUUCGAGAGCGCUGUUGCU________UGCUGUUUGCACUUUCGCCCAAAUGCCAACACAACCGAACAGCAACAGCGACAGAAAAA .........((.(.......).)).(((((((((.......(((((((((.......((......)).........))))))))).)))))))))..... (-15.49 = -16.87 + 1.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:24 2006