
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,295,136 – 14,295,269 |
| Length | 133 |
| Max. P | 0.963244 |

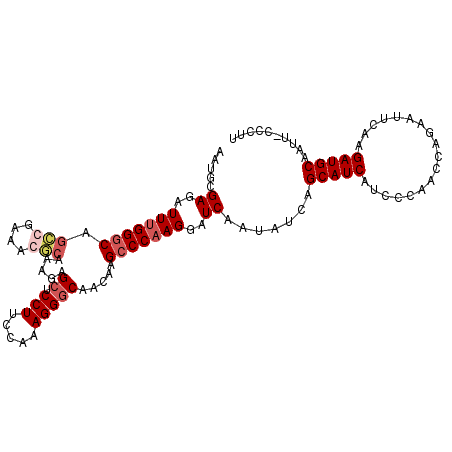
| Location | 14,295,136 – 14,295,240 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 87.34 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -22.58 |
| Energy contribution | -24.07 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14295136 104 + 22224390 UUUCGGAAUAAAAGUGUUGAAUGUAAAUGCGAGAUUUGGGCAGACGAAACUCAAAGAGCUCCUUCCAAAGGGCAACAGGCCCAAGGAUCAAUAUCAGCAUCAUC .............(((((((..((....))((..(((((((.((......)).....((.(((.....))))).....)))))))..))....))))))).... ( -24.90) >DroSec_CAF1 8021 104 + 1 CUUCGGAAUAAAAGUGUUGAAUGUGAAUGCGAGAUUUGGGCAGCCGAAAGGCAAAGAGCUCCUUCCAAAGGGCAACAAGCCCAAGGAUCAAUAUCAGCAUCAUC (....).......(((((((..((....))((..(((((((.(((....))).....((.(((.....))))).....)))))))..))....))))))).... ( -34.70) >DroSim_CAF1 8566 104 + 1 CUUCGGAAUAAAAGUGUUGAAUGUGAAUGCGAGAUUUGGGCAGCCGAAAGGCAAAGAGCUCCUUCCAAAGGGCAACAAGCCCAAGGAUCAAUAUCAGCAUCAUC (....).......(((((((..((....))((..(((((((.(((....))).....((.(((.....))))).....)))))))..))....))))))).... ( -34.70) >DroYak_CAF1 9783 97 + 1 ----AAAAUACAAGUGUUGAAUGUG---GCGAGAUUCGGCCAGCCGAAACUAGUAAAGCUCCUUCCAAAGGACAACGAGUCCAAGGAUCAAUAUCAGCAUCAUA ----.........(((((((..((.---((.((.(((((....))))).)).))...))(((((.....((((.....)))))))))......))))))).... ( -25.20) >consensus CUUCGGAAUAAAAGUGUUGAAUGUGAAUGCGAGAUUUGGGCAGCCGAAACGCAAAGAGCUCCUUCCAAAGGGCAACAAGCCCAAGGAUCAAUAUCAGCAUCAUC .............(((((((..((....))((..(((((((.(((....))).....((.(((.....))))).....)))))))..))....))))))).... (-22.58 = -24.07 + 1.50)

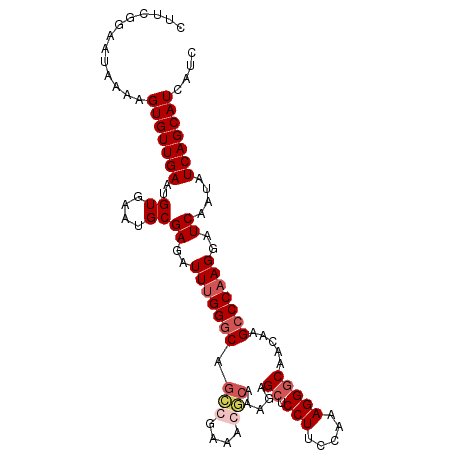

| Location | 14,295,161 – 14,295,269 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 91.40 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -21.77 |
| Energy contribution | -23.27 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14295161 108 + 22224390 AAUGCGAGAUUUGGGCAGACGAAACUCAAAGAGCUCCUUCCAAAGGGCAACAGGCCCAAGGAUCAAUAUCAGCAUCAUCCCAACCAGAAUUCAAGAUGCAAUU-CCCUU .....((..(((((((.((......)).....((.(((.....))))).....)))))))..)).......(((((..................)))))....-..... ( -24.07) >DroSec_CAF1 8046 108 + 1 AAUGCGAGAUUUGGGCAGCCGAAAGGCAAAGAGCUCCUUCCAAAGGGCAACAAGCCCAAGGAUCAAUAUCAGCAUCAUCCCAACCAGAAUUCAAGAUGCAAUU-CCCUU .....((..(((((((.(((....))).....((.(((.....))))).....)))))))..)).......(((((..................)))))....-..... ( -33.37) >DroSim_CAF1 8591 108 + 1 AAUGCGAGAUUUGGGCAGCCGAAAGGCAAAGAGCUCCUUCCAAAGGGCAACAAGCCCAAGGAUCAAUAUCAGCAUCAUCCCAACCAGAAUUCAAGAUGCAAUU-CCCUU .....((..(((((((.(((....))).....((.(((.....))))).....)))))))..)).......(((((..................)))))....-..... ( -33.37) >DroYak_CAF1 9804 106 + 1 ---GCGAGAUUCGGCCAGCCGAAACUAGUAAAGCUCCUUCCAAAGGACAACGAGUCCAAGGAUCAAUAUCAGCAUCAUACCAACCAGAAUUCAAGAUGCAAUUACCCUU ---...((.(((((....))))).)).((((.(.(((((.....((((.....))))))))).).......(((((..................)))))..)))).... ( -22.87) >consensus AAUGCGAGAUUUGGGCAGCCGAAACGCAAAGAGCUCCUUCCAAAGGGCAACAAGCCCAAGGAUCAAUAUCAGCAUCAUCCCAACCAGAAUUCAAGAUGCAAUU_CCCUU .....((..(((((((.(((....))).....((.(((.....))))).....)))))))..)).......(((((..................))))).......... (-21.77 = -23.27 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:02 2006