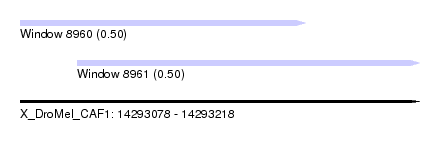
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,293,078 – 14,293,218 |
| Length | 140 |
| Max. P | 0.500000 |

| Location | 14,293,078 – 14,293,178 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.75 |
| Mean single sequence MFE | -37.07 |
| Consensus MFE | -26.80 |
| Energy contribution | -28.72 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
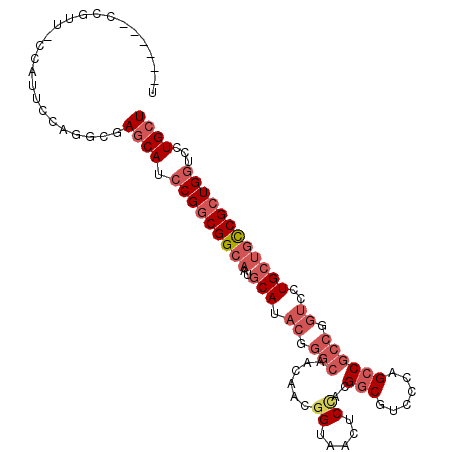
>X_DroMel_CAF1 14293078 100 + 22224390 U------CCGUU-UCGUUCUAGGCGAGCAUCCGGCGGCAAUGCAUACGGGCAACAACGGUAACUCUACGGCGUCCCAGCCGCCGGUCUUGCUGCCGCUGGUCCUGCU .------.....-((((.....))))(((.(((((((((..(((.((.(((.......(((....)))(((......)))))).))..))))))))))))...))). ( -41.10) >DroPse_CAF1 8990 94 + 1 UCCGAU---AUU-CAAUUGCAGGCGAACAUCCGGCGGCCAUGCAU---------CAUGGAAACUCGGCGCCACUUCAGCCACCGGCUUUGAUCUCGAUGUUGAUGCU ......---...-.....(((..(.((((((.((((.(((((...---------)))(....)..)))))).....((((...))))........))))))).))). ( -22.40) >DroSec_CAF1 6003 100 + 1 U------CCGUU-CCGUUUCAGGCGAGCAUCCGGCGGCAAUGCAUACGGGCAACAACGGUAACUCCACGGCGUCCCAGCCGCCGGUCCUGCUGCCGCUGGUCCUGCU .------.((((-........))))((((.(((((((((..(((.((.(((......((.....))..(((......)))))).))..))))))))))))...)))) ( -40.50) >DroSim_CAF1 6521 106 + 1 UCCGUUUCCGUU-UCGUUCCAGGCGAGCAUCCGGCGGCAAUGCAUACGGGCAACAACGGUAACUCCACGGCGUCCCAGCCGCCGGUCCUGCUGCCGCUGGUCCUGCU ............-((((.....))))(((.(((((((((..(((.((.(((......((.....))..(((......)))))).))..))))))))))))...))). ( -41.30) >DroEre_CAF1 5273 94 + 1 U-------------CAUUCCAGGCGAGCAUCCGGCGGCAAUGCACACGGGCAACAAUGGCAACUCCACGGCGAGCCAGCCGCCAGUCCUGCUGCCGCUGGUCCUGCU .-------------...........((((.(((((((((..(((...((((.....(((.....))).((((.(....))))).))))))))))))))))...)))) ( -37.30) >DroYak_CAF1 7652 107 + 1 UCCGUUUUCGUUUCCAUUCCAGGCGAGCAUCCGGCGGCAAUGCAUACGGGCAACAACGGUAACUCCACGGCGACCCAGCCGCCAGUCCUGCUGCCGCUGGUCCUGCU .......((((((.......))))))(((.(((((((((..(((.((.(((......((.....))..(((......)))))).))..))))))))))))...))). ( -39.80) >consensus U______CCGUU_CCAUUCCAGGCGAGCAUCCGGCGGCAAUGCAUACGGGCAACAACGGUAACUCCACGGCGUCCCAGCCGCCGGUCCUGCUGCCGCUGGUCCUGCU .........................((((.(((((((((..(((.((.(((......((.....))..(((......)))))).))..))))))))))))...)))) (-26.80 = -28.72 + 1.92)


| Location | 14,293,098 – 14,293,218 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.24 |
| Mean single sequence MFE | -44.38 |
| Consensus MFE | -21.32 |
| Energy contribution | -22.55 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.48 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14293098 120 + 22224390 CAUCCGGCGGCAAUGCAUACGGGCAACAACGGUAACUCUACGGCGUCCCAGCCGCCGGUCUUGCUGCCGCUGGUCCUGCUCACCUGCAGCACACUGAUGCUCCUCCAGCUGGUGGCCAGC ...(((((((((..(((.((.(((.......(((....)))(((......)))))).))..))))))))))))..(((..((((.((((((......))))......)).))))..))). ( -48.20) >DroPse_CAF1 9013 111 + 1 CAUCCGGCGGCCAUGCAU---------CAUGGAAACUCGGCGCCACUUCAGCCACCGGCUUUGAUCUCGAUGUUGAUGCUUGUCUGCAGCACUCUGAUUCUGCUGCACGCCUCUGCCUGC .....((((((((((...---------)))))......((((......(((.((.((((.(((....))).)))).)).)))..(((((((.........))))))))))).)))))... ( -33.60) >DroSim_CAF1 6547 120 + 1 CAUCCGGCGGCAAUGCAUACGGGCAACAACGGUAACUCCACGGCGUCCCAGCCGCCGGUCCUGCUGCCGCUGGUCCUGCUCACAUGCAGCACACUGAUGCUCCUCCAGCUGGUGGCCAGC ((((((((((((..(((.((.(((......((.....))..(((......)))))).))..)))))))))))((.((((......)))).))...))))........(((((...))))) ( -47.20) >DroEre_CAF1 5287 120 + 1 CAUCCGGCGGCAAUGCACACGGGCAACAAUGGCAACUCCACGGCGAGCCAGCCGCCAGUCCUGCUGCCGCUGGUCCUGCUCACCUGCAGCACGCUGAUGCUCCUCCAGCUGGUGGCCAGC .....(((((((.(((.((..(....)..))))).......((((.(....))))).....)))))))(((((((((((......))))((.((((.........))))))..))))))) ( -48.00) >DroWil_CAF1 5171 109 + 1 CAUCCGGCGGCAAUGCAU---------CAUGGCAACUCGGCGCCGCUGCAACCGCCGGCAAUGCUGUCAUUGCUCCUACUCACAUGCGGUGUACUGCUGCUAUUGCAAUCCUCCGCCC-- .....(((((...((((.---------.((((((.(...(((((((((........(((((((....)))))))........)).)))))))...).)))))))))).....))))).-- ( -39.79) >DroAna_CAF1 51670 111 + 1 CAUCCGGCGGCAAUGCA---------CCACGGCAACUCUGGGGCAUUUCGACCGCCGGGCCUGGUCUCGCUGCUGCUCUUCUUGUGCGGAGCGCCGCUGCUCAUCCAGGCGGUCACCUUU ...(((((((((((((.---------(((.(....)..))).)))))..).)))))))((((((....((.((.((.((((......)))).)).)).))....)))))).......... ( -49.50) >consensus CAUCCGGCGGCAAUGCAU________CAACGGCAACUCCACGGCGUUCCAGCCGCCGGUCCUGCUGCCGCUGGUCCUGCUCACCUGCAGCACACUGAUGCUCCUCCAGCCGGUGGCCAGC ...((((((((...................(....).....((....)).))))))))....((..(((((((....((.......(((....)))..))....)))).)))..)).... (-21.32 = -22.55 + 1.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:00 2006