
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,292,406 – 14,292,515 |
| Length | 109 |
| Max. P | 0.998432 |

| Location | 14,292,406 – 14,292,515 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.89 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.76 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
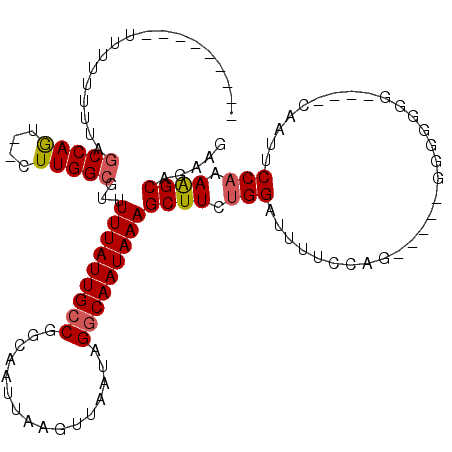
>X_DroMel_CAF1 14292406 109 + 22224390 ---------UUUUUUUUAGCCAGU--CUUGGCGUUUUUAUUGCCGGCAAUUAAGUUAAUAGGCAAUAAAGCUUCUGGAUUUUCCAGGGGGAGGGGGGGUUGGCAAUUCCAAAAGCAGAAG ---------.........((((..--(((..(..((((((((((................))))))))))(((((((.....)))))))..)..)))..))))................. ( -31.09) >DroPse_CAF1 8351 102 + 1 UCAGUGUUUUU-UUUUUAGCCAGUAGCUUGGCGUUUUUAUUGCCGGCAAUUAAGUUAAUAGGCAAUAAAGCUUCUGGAUUUGCCAGC---------------CAUCGCCAGAAGCAGU-- ...........-......(((((....)))))...(((((((((................)))))))))((((((((...((.....---------------))...))))))))...-- ( -30.09) >DroSec_CAF1 5355 99 + 1 ---------UUU-UUUUAGCCAGU--CUUGGCGUUUUUAUUGCCGGCAAUUAAGUUAAUAGACAAUAAAGCUUCUGGAUUUUCCA------GGGGGGG---CCAAUUCCAAAAGCAGAAG ---------...-.....(((...--...)))((((((......(((......(((....))).......(((((((.....)))------))))..)---))......))))))..... ( -23.10) >DroSim_CAF1 5851 100 + 1 ---------UU--UUUUACCCAAU--CUUGGCGUUUUUAUUGCCGGCAAUUAAGUUAAAAGGCAAUAAAGCUUCUGGAUUUUCCAG----AGGGGGGG---UCAAUUCCUAAGGCAGAAU ---------..--.....(((...--........((((((((((................))))))))))(((((((.....))))----)))))).(---((.........)))..... ( -26.79) >DroPer_CAF1 7915 105 + 1 UCAGUGUUUUUUUUUUUAGCCAGUAGCUUGGCGUUUUUAUUGCCGGCAAUUAAGUUAAUAGGCAAUAAAGCUUCUGGAUUUGCCAGC---------------CAUCGCCAGAAGCAGUGC ..................(((((....)))))...(((((((((................)))))))))((((((((...((.....---------------))...))))))))..... ( -30.09) >consensus _________UUUUUUUUAGCCAGU__CUUGGCGUUUUUAUUGCCGGCAAUUAAGUUAAUAGGCAAUAAAGCUUCUGGAUUUUCCAG_____GGGGGGG____CAAUUCCAAAAGCAGAAG ..................(((((....)))))...(((((((((................)))))))))((((.(((..............................))).))))..... (-18.48 = -18.76 + 0.28)


| Location | 14,292,406 – 14,292,515 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.89 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -20.80 |
| Energy contribution | -21.00 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.80 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14292406 109 - 22224390 CUUCUGCUUUUGGAAUUGCCAACCCCCCCUCCCCCUGGAAAAUCCAGAAGCUUUAUUGCCUAUUAACUUAAUUGCCGGCAAUAAAAACGCCAAG--ACUGGCUAAAAAAAA--------- .....(((((((((.((.(((..............))).)).)))))))))(((((((((................)))))))))...((((..--..)))).........--------- ( -28.33) >DroPse_CAF1 8351 102 - 1 --ACUGCUUCUGGCGAUG---------------GCUGGCAAAUCCAGAAGCUUUAUUGCCUAUUAACUUAAUUGCCGGCAAUAAAAACGCCAAGCUACUGGCUAAAAA-AAAAACACUGA --...((((((((...((---------------.....))...))))))))(((((((((................)))))))))...((((......))))......-........... ( -26.79) >DroSec_CAF1 5355 99 - 1 CUUCUGCUUUUGGAAUUGG---CCCCCCC------UGGAAAAUCCAGAAGCUUUAUUGUCUAUUAACUUAAUUGCCGGCAAUAAAAACGCCAAG--ACUGGCUAAAA-AAA--------- .....(((((((((.((..---((.....------.))..)))))))))))(((((((((................)))))))))...((((..--..)))).....-...--------- ( -25.69) >DroSim_CAF1 5851 100 - 1 AUUCUGCCUUAGGAAUUGA---CCCCCCCU----CUGGAAAAUCCAGAAGCUUUAUUGCCUUUUAACUUAAUUGCCGGCAAUAAAAACGCCAAG--AUUGGGUAAAA--AA--------- (((((......)))))..(---(((....(----((((.....))))).(((((((((((................)))))))))...))....--...))))....--..--------- ( -22.89) >DroPer_CAF1 7915 105 - 1 GCACUGCUUCUGGCGAUG---------------GCUGGCAAAUCCAGAAGCUUUAUUGCCUAUUAACUUAAUUGCCGGCAAUAAAAACGCCAAGCUACUGGCUAAAAAAAAAAACACUGA .....((((((((...((---------------.....))...))))))))(((((((((................)))))))))...((((......)))).................. ( -26.79) >consensus CUUCUGCUUUUGGAAUUG____CCCCCCC_____CUGGAAAAUCCAGAAGCUUUAUUGCCUAUUAACUUAAUUGCCGGCAAUAAAAACGCCAAG__ACUGGCUAAAAAAAA_________ .....((((((((..............................))))))))(((((((((................)))))))))...((((......)))).................. (-20.80 = -21.00 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:58 2006