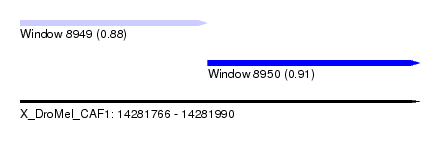
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,281,766 – 14,281,990 |
| Length | 224 |
| Max. P | 0.912832 |

| Location | 14,281,766 – 14,281,871 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 86.38 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -20.25 |
| Energy contribution | -19.94 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14281766 105 + 22224390 GAUCGAAUCCAUCGCUUAUACAUCCAGUGCAUUCAAAUA-CAUGUAUGUAUAAUGUGUAUGCAUGGAGUUCGGGUGCGUGUGUGCGGUGCAUAUUGAAUUUAUAAU ..((((((.((((((.(((((.((((.(((((.((.(((-((....)))))....)).)))))))))((......))))))).)))))).)).))))......... ( -28.50) >DroSec_CAF1 22269 102 + 1 GAUCGAAUCCAUCGCUUAUACAUCCAGUGC---CACAUA-CAUGUACAUAUAAUAUGUAUGCAUGGAGUUGGGGUGCGUGUGUGCGGUGCAUAUUGAAUUUAUAAU ..((((((.((((((.(((((.(((((..(---((((((-(((((.......)))))))))..)))..)))))....))))).)))))).)).))))......... ( -31.60) >DroSim_CAF1 26351 102 + 1 GAUCGAAUCCAUCGCUUAUACAUCCAGUGC---CACAUA-CAUGUAUAUAUAAUAUGUAUGCAUGGAGUUGGGGUGCGUGUGUGCGGUGCAUAUUGAAUUUAUAAU ..((((((.((((((.(((((.(((((..(---((((((-(((((.......)))))))))..)))..)))))....))))).)))))).)).))))......... ( -31.60) >DroEre_CAF1 24254 89 + 1 GAUCGAGUCCAUCGCUUAU---UCCAGUGC---CAUGUAU-----------AAUAUGUAUGCAUGGAGCUCGGGCGCGUGUGUGCGAUGCAUAUUGAAUUUAUAAU ..((((...((((((....---.(((((.(---(((((((-----------(.....))))))))).))).))(((....)))))))))....))))......... ( -27.50) >consensus GAUCGAAUCCAUCGCUUAUACAUCCAGUGC___CACAUA_CAUGUAUAUAUAAUAUGUAUGCAUGGAGUUCGGGUGCGUGUGUGCGGUGCAUAUUGAAUUUAUAAU ..((((((.((((((.(((((.((((.(((....(((((..............)))))..)))))))((......))))))).)))))).)).))))......... (-20.25 = -19.94 + -0.31)


| Location | 14,281,871 – 14,281,990 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.82 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -37.18 |
| Energy contribution | -37.46 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14281871 119 + 22224390 GCACGGUUGAUAGAUUCUGGCUUUGCAUCCUGUCAACUGCAAGCUGUUUGCCAGGACUGCCUUUUGUCCCGAUUGUUCGGACAGGCGGAGUCAAUCAACGGACAAAAGUUGAUUUCGCA- ((...((((((.(((((((.(...((((((((.(((..((.....))))).))))).)))....(((((.((....)))))))).))))))).)))))).(((....)))......)).- ( -39.00) >DroSec_CAF1 22371 120 + 1 GCACGGUUGAUAGAUUCUGGCUUUGCAUCCUGUCAACUGCAAGCUGUUUGCCACGACUGCCUUUUGUCCCGAUUGUUCGGACAGGCGGGGUCAAUCAACGGACAAAAGUCGAUUUCGCAA ((...((((((.(((((((((...(((.(.(((.....))).).)))..)))).))((((((...((((.((....)))))))))))).))).)))))).(((....)))......)).. ( -37.50) >DroSim_CAF1 26453 119 + 1 GCACGGUUGAUAGAUUCUGGCUUUGCAUCCUGUCAACUGCAAGCUGUUUGCCAGGACUGCCUUUUGUCCCGAUUGUUCGGACAGGCGGAGUCAAUCAACGGACAAAAGUCGAUUUCGCA- ((...((((((.(((((((.(...((((((((.(((..((.....))))).))))).)))....(((((.((....)))))))).))))))).)))))).(((....)))......)).- ( -41.20) >DroEre_CAF1 24343 119 + 1 GCACGCUUGAUAGAUUCUGGCUUUGCAUCCUGUCAACUGCAAGCUGUUUGCCACGACUGCCUUUUGUCCCGAUUGUUCGGACAGGCGGAGUCAAUCAACGGACAAAAGUCGAUUUCGCA- ((.((.(((((.(((((((((...(((.(.(((.....))).).)))..)))......((((...((((.((....)))))))))))))))).)))))))(((....)))......)).- ( -36.90) >DroYak_CAF1 25873 119 + 1 GCACGGUUGAUAGAUUCUGGCUUUGCAUCCUGUCAACUGCAAGCUGUUUGCCAGUACUGCCUUUUGUCCCGAUUGUUCGGACAGGCGGAGUCAAUCAACGGACAAAAGUCGAUUUCGCA- ((...((((((.(((((((((...(((.(.(((.....))).).)))..)))))..((((((...((((.((....)))))))))))))))).)))))).(((....)))......)).- ( -39.80) >consensus GCACGGUUGAUAGAUUCUGGCUUUGCAUCCUGUCAACUGCAAGCUGUUUGCCAGGACUGCCUUUUGUCCCGAUUGUUCGGACAGGCGGAGUCAAUCAACGGACAAAAGUCGAUUUCGCA_ ((...((((((.(((((((((...(((.(.(((.....))).).)))..)))))..((((((...((((.((....)))))))))))))))).)))))).(((....)))......)).. (-37.18 = -37.46 + 0.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:50 2006