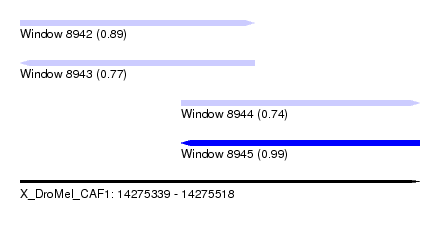
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,275,339 – 14,275,518 |
| Length | 179 |
| Max. P | 0.987715 |

| Location | 14,275,339 – 14,275,444 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -21.30 |
| Energy contribution | -20.98 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14275339 105 + 22224390 AAAUCCGGCUGAAUUCAAAAUGCAAUCAAAUUUAUGCAUAACAAUAUUUGGCGAGCUCAAGGAUUGGCAA-AG--------------UAUGUGAGUGAGUUUUCGCCAAUUACAGUGGAG ...((((.(((........(((((..........))))).....((.((((((((((((....((.(((.-..--------------..))).)))))))..))))))).))))))))). ( -26.50) >DroSec_CAF1 15976 106 + 1 GAAUCCGGCUGAAAUCAAAAUGCAAUCAAAUUUAUGUAUAACAAUAUUUGGCGAGCUCAAGGAUUGGCAAAAG--------------UAUGUGAGUGAGUUUUCGCCAAUUACAGUGGAG ...((((.(((........(((((..........))))).....((.((((((((((((.......((....)--------------).......)))))..))))))).))))))))). ( -25.14) >DroSim_CAF1 19480 106 + 1 GAAUCCGGCUGAAAUCAAAAUGCAAUCAAAUUUAUGCAUAACAAUAUUUGGCGAGCUCAAGGAUUGGCAAAAG--------------UAUGUGAGUGAGUUUUCGCCAAUUACAGUGGAG ...((((.(((........(((((..........))))).....((.((((((((((((.......((....)--------------).......)))))..))))))).))))))))). ( -27.14) >DroEre_CAF1 18390 119 + 1 GAAUCCGACUGAAAUCAAAAUGCAAUCAAAUUUAUGCAUAACAAUAUUUGGCGAGCUCAAGGAUUGCGAA-CGUACAUAUGUACGUGUAUGUGAGUGAGUUUUCGCCAAUUACUGUGGAG ...(((.............(((((..........))))).(((.((.((((((((((((....(..((.(-(((((....))))))...))..).)))))..))))))).)).)))))). ( -35.70) >DroYak_CAF1 19178 105 + 1 GAAUCCGACUGAAAUCAAAAUGCAAUCAAAUUUAUGCAUAACAAUAUUUGGCGAGCUCGAGGAUUGGGAA-AG--------------UAUGAGAGUGAGUUUUCGCCAAUUAUUGUGGAG ...(((.............(((((..........))))).((((((.((((((((((((....(((....-..--------------..)))...)))))..))))))).))))))))). ( -26.80) >consensus GAAUCCGGCUGAAAUCAAAAUGCAAUCAAAUUUAUGCAUAACAAUAUUUGGCGAGCUCAAGGAUUGGCAA_AG______________UAUGUGAGUGAGUUUUCGCCAAUUACAGUGGAG ...(((.((((........(((((..........))))).....((.((((((((((((....................................)))))..))))))).))))))))). (-21.30 = -20.98 + -0.32)



| Location | 14,275,339 – 14,275,444 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -22.06 |
| Consensus MFE | -16.60 |
| Energy contribution | -16.64 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14275339 105 - 22224390 CUCCACUGUAAUUGGCGAAAACUCACUCACAUA--------------CU-UUGCCAAUCCUUGAGCUCGCCAAAUAUUGUUAUGCAUAAAUUUGAUUGCAUUUUGAAUUCAGCCGGAUUU .(((.(((...(((((((...((((........--------------..-...........)))).)))))))...(((..(((((..........)))))..)))...)))..)))... ( -19.80) >DroSec_CAF1 15976 106 - 1 CUCCACUGUAAUUGGCGAAAACUCACUCACAUA--------------CUUUUGCCAAUCCUUGAGCUCGCCAAAUAUUGUUAUACAUAAAUUUGAUUGCAUUUUGAUUUCAGCCGGAUUC .(((.(((..((((((((((.............--------------.))))))))))......((....(((((..((.....))...)))))...))..........)))..)))... ( -17.04) >DroSim_CAF1 19480 106 - 1 CUCCACUGUAAUUGGCGAAAACUCACUCACAUA--------------CUUUUGCCAAUCCUUGAGCUCGCCAAAUAUUGUUAUGCAUAAAUUUGAUUGCAUUUUGAUUUCAGCCGGAUUC .(((.(((...(((((((...((((........--------------..............)))).)))))))..((((..(((((..........)))))..))))..)))..)))... ( -20.05) >DroEre_CAF1 18390 119 - 1 CUCCACAGUAAUUGGCGAAAACUCACUCACAUACACGUACAUAUGUACG-UUCGCAAUCCUUGAGCUCGCCAAAUAUUGUUAUGCAUAAAUUUGAUUGCAUUUUGAUUUCAGUCGGAUUC .(((((((((.(((((((...((((.........((((((....)))))-)..........)))).))))))).)))))).(((((..........)))))...(((....))))))... ( -31.41) >DroYak_CAF1 19178 105 - 1 CUCCACAAUAAUUGGCGAAAACUCACUCUCAUA--------------CU-UUCCCAAUCCUCGAGCUCGCCAAAUAUUGUUAUGCAUAAAUUUGAUUGCAUUUUGAUUUCAGUCGGAUUC .(((((((((.(((((((...(((.........--------------..-............))).))))))).)))))).(((((..........)))))...(((....))))))... ( -22.00) >consensus CUCCACUGUAAUUGGCGAAAACUCACUCACAUA______________CU_UUGCCAAUCCUUGAGCUCGCCAAAUAUUGUUAUGCAUAAAUUUGAUUGCAUUUUGAUUUCAGCCGGAUUC .(((((.(((.(((((((...(((......................................))).))))))).))).)).(((((..........))))).............)))... (-16.60 = -16.64 + 0.04)


| Location | 14,275,411 – 14,275,518 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -20.23 |
| Energy contribution | -20.97 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14275411 107 + 22224390 UAUGUGAGUGAGUUUUCGCCAAUUACAGUGGAGAACAACUGAAAGGAUAGAGAUUUUGAGAAAAUGUAAGCCUUGAAUGGCUUGUAUUUGGU------GUAAUUCACUUUGAG .....(((((((((..((((.....((((........))))....................(((((((((((......))))))))))))))------).))))))))).... ( -29.30) >DroSec_CAF1 16049 107 + 1 UAUGUGAGUGAGUUUUCGCCAAUUACAGUGGAGAACAACUGAAAGGAUAGAGAUUUUGAGAAAAUGUAAGCCUUGAAUGGCUUGUAUUUGAU------GUAUUUCACUUUGAG ...(((((......)))))...(((.(((((((.(((.((.((((........)))).)).(((((((((((......)))))))))))..)------)).))))))).))). ( -25.20) >DroSim_CAF1 19553 107 + 1 UAUGUGAGUGAGUUUUCGCCAAUUACAGUGGAGAACAACUGAAAGGAUAGAGAUUUUGAGAAAGUGUAAGCCUUGAAUGGCUUGUAUUUGAU------GUAUUUCACUUUGAG ...(((((......)))))...(((.(((((((.(((.((.((((........)))).)).(((((((((((......)))))))))))..)------)).))))))).))). ( -25.20) >DroYak_CAF1 19250 109 + 1 UAUGAGAGUGAGUUUUCGCCAAUUAUUGUGGAGAACAACUGAAAGGAUAGAGAUUCGGCAAAGUUGUAAGCCUUGAAUGGCUU----UUGGUGUACGAGUAUUUCACUUUGAG ..(.((((((((..((((((((............((((((....((((....)))).....))))))(((((......)))))----))))....))))...)))))))).). ( -27.40) >consensus UAUGUGAGUGAGUUUUCGCCAAUUACAGUGGAGAACAACUGAAAGGAUAGAGAUUUUGAGAAAAUGUAAGCCUUGAAUGGCUUGUAUUUGAU______GUAUUUCACUUUGAG .....(((((((.((((.((.....((((........))))...))...))))........(((((((((((......))))))))))).............))))))).... (-20.23 = -20.97 + 0.75)


| Location | 14,275,411 – 14,275,518 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -17.12 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.33 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14275411 107 - 22224390 CUCAAAGUGAAUUAC------ACCAAAUACAAGCCAUUCAAGGCUUACAUUUUCUCAAAAUCUCUAUCCUUUCAGUUGUUCUCCACUGUAAUUGGCGAAAACUCACUCACAUA .....(((((....(------.((((.((((((((......)))))...........................(((.(....).)))))).)))).).....)))))...... ( -16.60) >DroSec_CAF1 16049 107 - 1 CUCAAAGUGAAAUAC------AUCAAAUACAAGCCAUUCAAGGCUUACAUUUUCUCAAAAUCUCUAUCCUUUCAGUUGUUCUCCACUGUAAUUGGCGAAAACUCACUCACAUA ......((((.....------.........(((((......)))))...........................((((.(((.(((.......))).)))))))...))))... ( -15.90) >DroSim_CAF1 19553 107 - 1 CUCAAAGUGAAAUAC------AUCAAAUACAAGCCAUUCAAGGCUUACACUUUCUCAAAAUCUCUAUCCUUUCAGUUGUUCUCCACUGUAAUUGGCGAAAACUCACUCACAUA ......((((.....------.........(((((......)))))...........................((((.(((.(((.......))).)))))))...))))... ( -15.90) >DroYak_CAF1 19250 109 - 1 CUCAAAGUGAAAUACUCGUACACCAA----AAGCCAUUCAAGGCUUACAACUUUGCCGAAUCUCUAUCCUUUCAGUUGUUCUCCACAAUAAUUGGCGAAAACUCACUCUCAUA .....(((((................----(((((......))))).....(((((((((..........))).(((((.....)))))....))))))...)))))...... ( -20.10) >consensus CUCAAAGUGAAAUAC______ACCAAAUACAAGCCAUUCAAGGCUUACAUUUUCUCAAAAUCUCUAUCCUUUCAGUUGUUCUCCACUGUAAUUGGCGAAAACUCACUCACAUA .....(((((....................(((((......)))))............................(((.(((.(((.......))).)))))))))))...... (-15.32 = -15.33 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:46 2006