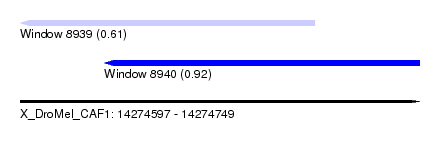
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,274,597 – 14,274,749 |
| Length | 152 |
| Max. P | 0.915867 |

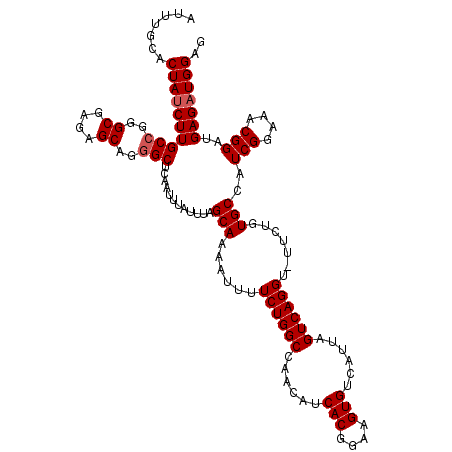
| Location | 14,274,597 – 14,274,709 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.21 |
| Mean single sequence MFE | -34.24 |
| Consensus MFE | -21.24 |
| Energy contribution | -22.24 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14274597 112 - 22224390 UUUAGCAAAAUUUUCUGGCCAACAUCACGGAAGUGUCAUUAGUCAGGUUUUCUGUGCCAUCGGAAACGGAUGAGCUGGAGAAUCCUGGCGAGAAUUGGG-GGAAUGGAUCCAU------- .....(..(((((((..........(((....)))......(((((((((((((.(((((((....).)))).))))))))).)))))))))))))..)-(((.....)))..------- ( -36.30) >DroSec_CAF1 15254 92 - 1 UUUAGCAAAAUUUUCUGGCCAACAUCACGGAAGUGUCAUUAGUCAGGU-UUCUGUGCCAUCGGAAACGGAUGAGAUGGAGA--------------------GAAUGAAGCUAU------- ..((((...(((((((.........(((....)))(((((.(((..((-(((((......))))))).)))..))))))))--------------------))))...)))).------- ( -27.50) >DroSim_CAF1 18738 112 - 1 UUUAGCAAAAUUUUCUGGCCAACAUCACGGAAGUGUCAUUAGUCAGGU-UUCUGUGCCAUCGGAAACGGAUGAGAUGGAGAAUCCUGGCGAGAAUUGGGGGGGAUGAAGCUAU------- ..((((...(((..((..((((...(((....)))......(((((((-((((((..(((((....).))))..)))))))..)))))).....))))))..)))...)))).------- ( -37.20) >DroEre_CAF1 17684 110 - 1 UUUUGCACAAUUUUCUGGCCAACAUCACGGAAGUGUCAUUAGUCAGGU-UUCUGUGCCAUCGGAAACGGAUGAGAUGGAGAAUCCGUGGGGGGAUCGGG-UUAAUGCAGCUU-------- ..(((((......(((((((....(((((((....(((((.(((..((-(((((......))))))).)))..)))))....)))))))..)).)))))-....)))))...-------- ( -35.00) >DroYak_CAF1 18466 118 - 1 UUUAGCAAAAUUUUCUGGCCAACAUCACGGAAGUGUCAUUAGUCAGGU-UUCUGUGCCAUCGGAAACGGAUGAGAUGGAGAAUCCUGCGGAGAAUUGGG-GGAAUGCAGCUAUGCAGCUA ..((((..(((((((((........(((....)))........(((((-((((((..(((((....).))))..)))))))..)))))))))))))...-....((((....)))))))) ( -35.20) >consensus UUUAGCAAAAUUUUCUGGCCAACAUCACGGAAGUGUCAUUAGUCAGGU_UUCUGUGCCAUCGGAAACGGAUGAGAUGGAGAAUCCUGGCGAGAAUUGGG_GGAAUGAAGCUAU_______ ..((((.......((((((......(((....)))......))))))..((((((..(((((....).))))..))))))............................))))........ (-21.24 = -22.24 + 1.00)

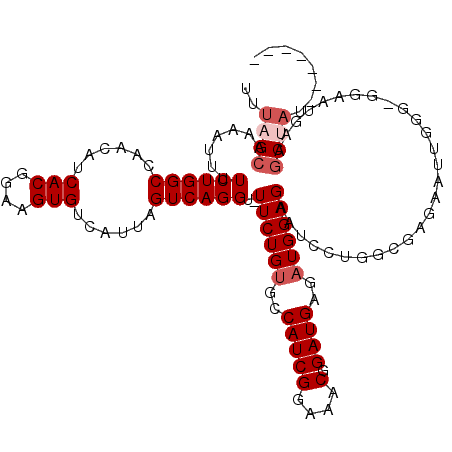
| Location | 14,274,629 – 14,274,749 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.65 |
| Mean single sequence MFE | -36.75 |
| Consensus MFE | -33.92 |
| Energy contribution | -34.32 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14274629 120 - 22224390 AUUUGCACUAUCUUGCCGGGCGAAAGCAGGGCUCAAUUUAUUUAGCAAAAUUUUCUGGCCAACAUCACGGAAGUGUCAUUAGUCAGGUUUUCUGUGCCAUCGGAAACGGAUGAGCUGGAG .(((((........(((..((....))..)))............)))))....((((((......(((....)))......))))))...((((.(((((((....).)))).)))))). ( -34.85) >DroSec_CAF1 15267 119 - 1 AUUUUCACUAUCUUGCAGGGCGAGAGCAGGGCUCAAUUUAUUUAGCAAAAUUUUCUGGCCAACAUCACGGAAGUGUCAUUAGUCAGGU-UUCUGUGCCAUCGGAAACGGAUGAGAUGGAG .......(((((((....(((..((((...))))..........(((.((...((((((......(((....)))......)))))).-)).)))))).(((....)))..))))))).. ( -34.50) >DroSim_CAF1 18771 119 - 1 AUUUGCACUAUCUUGCCGGGCGAGAGCAGGGCUCAAUUUAUUUAGCAAAAUUUUCUGGCCAACAUCACGGAAGUGUCAUUAGUCAGGU-UUCUGUGCCAUCGGAAACGGAUGAGAUGGAG .......((((((((((..((....))..)))............(((......((((((......(((....)))......)))))).-.....)))..(((....)))..))))))).. ( -37.00) >DroEre_CAF1 17715 119 - 1 AUUUGCACUAUCUUGCCGGGCGAGAGCAGGGCUCAAUUUAUUUUGCACAAUUUUCUGGCCAACAUCACGGAAGUGUCAUUAGUCAGGU-UUCUGUGCCAUCGGAAACGGAUGAGAUGGAG .......((((((((((..((....))..)))............(((((....((((((......(((....)))......)))))).-...)))))..(((....)))..))))))).. ( -40.40) >DroYak_CAF1 18505 119 - 1 AUUUGCACUAUCUUGCCGGGCGAGAGCAGGGCUCAAUUUAUUUAGCAAAAUUUUCUGGCCAACAUCACGGAAGUGUCAUUAGUCAGGU-UUCUGUGCCAUCGGAAACGGAUGAGAUGGAG .......((((((((((..((....))..)))............(((......((((((......(((....)))......)))))).-.....)))..(((....)))..))))))).. ( -37.00) >consensus AUUUGCACUAUCUUGCCGGGCGAGAGCAGGGCUCAAUUUAUUUAGCAAAAUUUUCUGGCCAACAUCACGGAAGUGUCAUUAGUCAGGU_UUCUGUGCCAUCGGAAACGGAUGAGAUGGAG .......((((((((((..((....))..)))............(((......((((((......(((....)))......)))))).......)))..(((....)))..))))))).. (-33.92 = -34.32 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:41 2006