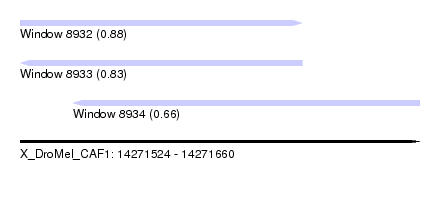
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,271,524 – 14,271,660 |
| Length | 136 |
| Max. P | 0.879836 |

| Location | 14,271,524 – 14,271,620 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.10 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -16.64 |
| Energy contribution | -17.40 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14271524 96 + 22224390 CCGUAGU---------------------UUUGUUGCUGC-CGUGACA-GGAGACAGCUACCGCAGCAGCUGAAAAUUCGAGCUAGUACACUGCGAGAAAA-AGAGCUGAUGCAUGCAAAU ..(((((---------------------(.(((..(...-.)..)))-(....))))))).(((.(((((.....(((..((.((....))))..)))..-..))))).)))........ ( -27.20) >DroSec_CAF1 12119 93 + 1 CCGUAGU---------------------UUUGUUGCUGC-AGUGACA-GGAGACAGCUACCGCAGCAGCUGAAAAUUCCAGCUAGUACACUGCGAGAAAA-AAAGCAGAUGCCAGCA--- ..(.(((---------------------(((((((((((-.(((.(.-(....).).))).))))))))..)))))).).(((.(((..((((.......-...)))).))).))).--- ( -29.10) >DroSim_CAF1 16148 95 + 1 CCGUAGU---------------------UUUGUUGCUGC-AGUGACA-GGAGACAGCUACCGCAGCAGCUGAAAAUUCCAGCUAGUACACUGCGAGAAA--AAAGUAGAUGCCAACAAGA .((((((---------------------......(((((-.(((.(.-(....).).))).)))))(((((.......))))).....)))))).....--................... ( -25.80) >DroEre_CAF1 14546 86 + 1 CUGUAGU------------------------UUUGUUGCAAGUGGCAAGGCGACAGCUACCGCAGCAGCUGAGCACUCGAACUAGUGCACUGCGAGAAAU-AAAGG---------CAAAU .((((((------------------------.(((((((..(((((..(....).))))).)))))))....(((((......)))))))))))......-.....---------..... ( -25.00) >DroYak_CAF1 15211 114 + 1 CCGCAGUUCUUUUUUUUUCUUAUUUUUUUUUUUUGCUGC-AGUGACA-GGAGACAGCUACCGCAGCAGCUGAAAACUCCAACUAGUACACUGCAAAAAAAGAAAAG----CGCUACAAGC .(((..(((((((((.....................(((-((((((.-((((.(((((........)))))....)))).....)).))))))))))))))))..)----))........ ( -28.59) >consensus CCGUAGU_____________________UUUGUUGCUGC_AGUGACA_GGAGACAGCUACCGCAGCAGCUGAAAAUUCCAGCUAGUACACUGCGAGAAAA_AAAGC_GAUGCCAACAAAU .((((((...........................(((((.(((.....(....).)))...)))))(((((.......))))).....)))))).......................... (-16.64 = -17.40 + 0.76)



| Location | 14,271,524 – 14,271,620 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.10 |
| Mean single sequence MFE | -27.76 |
| Consensus MFE | -16.28 |
| Energy contribution | -16.48 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14271524 96 - 22224390 AUUUGCAUGCAUCAGCUCU-UUUUCUCGCAGUGUACUAGCUCGAAUUUUCAGCUGCUGCGGUAGCUGUCUCC-UGUCACG-GCAGCAACAAA---------------------ACUACGG ...(((.((((.(((((..-.....(((.(((......))))))......))))).)))))))((((((...-......)-)))))......---------------------....... ( -25.02) >DroSec_CAF1 12119 93 - 1 ---UGCUGGCAUCUGCUUU-UUUUCUCGCAGUGUACUAGCUGGAAUUUUCAGCUGCUGCGGUAGCUGUCUCC-UGUCACU-GCAGCAACAAA---------------------ACUACGG ---..((((((.((((...-.......)))))))...(((((((...)))))))((((((((..(.......-.)..)))-)))))......---------------------....))) ( -29.10) >DroSim_CAF1 16148 95 - 1 UCUUGUUGGCAUCUACUUU--UUUCUCGCAGUGUACUAGCUGGAAUUUUCAGCUGCUGCGGUAGCUGUCUCC-UGUCACU-GCAGCAACAAA---------------------ACUACGG ..(((((((......)...--......((((((.((.....(((.....(((((((....)))))))..)))-.))))))-))..)))))).---------------------....... ( -25.90) >DroEre_CAF1 14546 86 - 1 AUUUG---------CCUUU-AUUUCUCGCAGUGCACUAGUUCGAGUGCUCAGCUGCUGCGGUAGCUGUCGCCUUGCCACUUGCAACAAA------------------------ACUACAG ..(((---------(....-.......))))((((..(((.((((.((.(((((((....)))))))..))))))..))))))).....------------------------....... ( -21.70) >DroYak_CAF1 15211 114 - 1 GCUUGUAGCG----CUUUUCUUUUUUUGCAGUGUACUAGUUGGAGUUUUCAGCUGCUGCGGUAGCUGUCUCC-UGUCACU-GCAGCAAAAAAAAAAAAUAAGAAAAAAAAAGAACUGCGG ........((----(..((((((((((((((((.((.....((((....(((((((....))))))).))))-.))))))-))).....................)))))))))..))). ( -37.09) >consensus ACUUGCAGGCAUC_CCUUU_UUUUCUCGCAGUGUACUAGCUGGAAUUUUCAGCUGCUGCGGUAGCUGUCUCC_UGUCACU_GCAGCAACAAA_____________________ACUACGG .........................(((((((.....((((((.....)))))))))))))..(((((.............))))).................................. (-16.28 = -16.48 + 0.20)

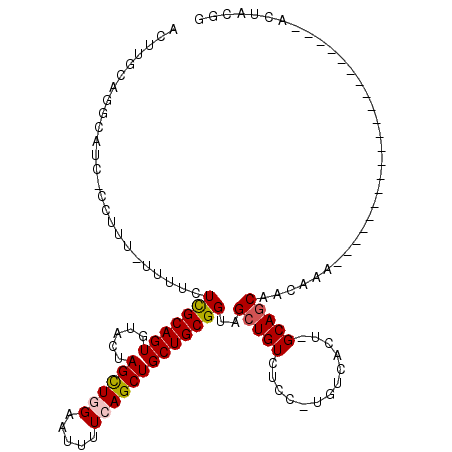
| Location | 14,271,542 – 14,271,660 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.50 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.655042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14271542 118 - 22224390 AACAUGGCAGCACGAUUUUAAAUCUAUAUUUUCUAUAACAAUUUGCAUGCAUCAGCUCU-UUUUCUCGCAGUGUACUAGCUCGAAUUUUCAGCUGCUGCGGUAGCUGUCUCCUGUCACG .(((.((((((..(((.....)))...................(((.((((.(((((..-.....(((.(((......))))))......))))).)))))))))))))...))).... ( -26.62) >DroSec_CAF1 12137 105 - 1 AACAAGGCACCACGAUUUGAAAUCUAUAUU-------------UGCUGGCAUCUGCUUU-UUUUCUCGCAGUGUACUAGCUGGAAUUUUCAGCUGCUGCGGUAGCUGUCUCCUGUCACU .....((((.....................-------------.(((((.((((((...-.......)))).)).))))).(((.....(((((((....)))))))..)))))))... ( -26.40) >DroSim_CAF1 16166 117 - 1 AACUUGGCACUACGAUCUGAAAUCUAUAUUCUCUAUAACAUCUUGUUGGCAUCUACUUU--UUUCUCGCAGUGUACUAGCUGGAAUUUUCAGCUGCUGCGGUAGCUGUCUCCUGUCACU .....((((((((.....((((.............((((.....))))...........--)))).((((((.....(((((((...))))))))))))))))).)))).......... ( -25.25) >DroYak_CAF1 15250 92 - 1 AACAAGGCAGCA--AUUC---------------------GGCUUGUAGCG----CUUUUCUUUUUUUGCAGUGUACUAGUUGGAGUUUUCAGCUGCUGCGGUAGCUGUCUCCUGUCACU .....(((((..--...(---------------------((((.(((.((----((.............))))))).)))))(((....(((((((....))))))).))))))))... ( -27.02) >consensus AACAAGGCACCACGAUUUGAAAUCUAUAUU_________A_CUUGCAGGCAUCUGCUUU_UUUUCUCGCAGUGUACUAGCUGGAAUUUUCAGCUGCUGCGGUAGCUGUCUCCUGUCACU .....((((((.................................((........)).........(((((((.....(((((((...))))))))))))))..)))))).......... (-19.20 = -19.20 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:36 2006