
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,271,311 – 14,271,511 |
| Length | 200 |
| Max. P | 0.999248 |

| Location | 14,271,311 – 14,271,414 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 94.29 |
| Mean single sequence MFE | -16.58 |
| Consensus MFE | -15.18 |
| Energy contribution | -15.38 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.999248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
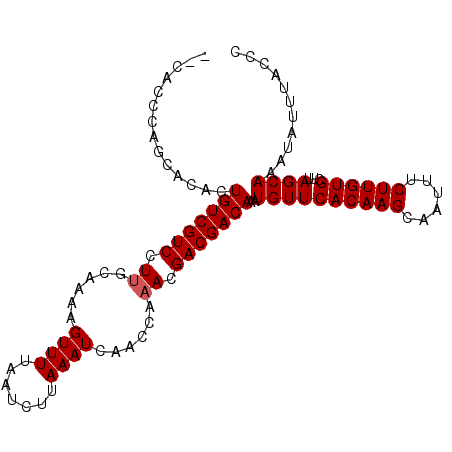
>X_DroMel_CAF1 14271311 103 + 22224390 AACACCCAACACCCUGUCGUCCUUGCAAAAGUUUUAAUCUUAAAUCAACCAAACGACGACAAAUGUUCACAAGCAAUUUCUUGUGCUUAGCAAAUAUUUACCC ..............(((((((.((......((((.......))))......)).)))))))..((((((((((......))))))...))))........... ( -15.60) >DroSec_CAF1 11912 101 + 1 --CACCCAUCACUCUGUCGUCCUUGCAAAAGUUUUAAUCUUAAAUCAACCAAACGACGACAAAUGUUCACAAGCAAUUUCUUGUGCUUAGCAAAUAUUUACCC --............(((((((.((......((((.......))))......)).)))))))..((((((((((......))))))...))))........... ( -15.60) >DroSim_CAF1 15938 101 + 1 --CACCCAGCACACUGUCGUCCUUGCAAAAGUUUUAAUCUUAAAUCAACCAAACGACGACAAAUGUUCACAAGCAAUUUCUUGUGCUUAGCAAAUAUUUACCC --......((....(((((((.((......((((.......))))......)).)))))))..((..((((((......))))))..))))............ ( -16.90) >DroEre_CAF1 14357 99 + 1 ----CACGGCAAACUGUCGUCAUGGCAAAAGUUUUAAUCUUAAAUCAACCAAACGACGACAAAUGUUCACAAGCAAUUUCUUGUGCUUAGCAAAUAUUUACCC ----....((....(((((((.(((.....((((.......))))...)))...)))))))..((..((((((......))))))..))))............ ( -17.90) >DroYak_CAF1 15004 99 + 1 ----CACAGCACACUGUCGUCAUUGCAAAAGUUUUAAUCUUAAAUCAACCAAACGACGACAAAUGUUCACAAGCAAUUUCUUGUGCUUAGCAAAUAUUUACCC ----....((....(((((((.((......((((.......))))......)).)))))))..((..((((((......))))))..))))............ ( -16.90) >consensus __CACCCAGCACACUGUCGUCCUUGCAAAAGUUUUAAUCUUAAAUCAACCAAACGACGACAAAUGUUCACAAGCAAUUUCUUGUGCUUAGCAAAUAUUUACCC ..............(((((((.((......((((.......))))......)).)))))))..((((((((((......))))))...))))........... (-15.18 = -15.38 + 0.20)


| Location | 14,271,311 – 14,271,414 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 94.29 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -20.22 |
| Energy contribution | -20.42 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14271311 103 - 22224390 GGGUAAAUAUUUGCUAAGCACAAGAAAUUGCUUGUGAACAUUUGUCGUCGUUUGGUUGAUUUAAGAUUAAAACUUUUGCAAGGACGACAGGGUGUUGGGUGUU .(((((....)))))...((((((......))))))(((((((((((((.(((((((.......))))))).(((....)))))))))).))))))....... ( -24.40) >DroSec_CAF1 11912 101 - 1 GGGUAAAUAUUUGCUAAGCACAAGAAAUUGCUUGUGAACAUUUGUCGUCGUUUGGUUGAUUUAAGAUUAAAACUUUUGCAAGGACGACAGAGUGAUGGGUG-- .(((((....)))))...((((((......)))))).((.(((((((((.(((((((.......))))))).(((....))))))))))))))........-- ( -22.80) >DroSim_CAF1 15938 101 - 1 GGGUAAAUAUUUGCUAAGCACAAGAAAUUGCUUGUGAACAUUUGUCGUCGUUUGGUUGAUUUAAGAUUAAAACUUUUGCAAGGACGACAGUGUGCUGGGUG-- ............(((.((((((((......)))))..(((((.((((((.(((((((.......))))))).(((....))))))))))))))))).))).-- ( -24.10) >DroEre_CAF1 14357 99 - 1 GGGUAAAUAUUUGCUAAGCACAAGAAAUUGCUUGUGAACAUUUGUCGUCGUUUGGUUGAUUUAAGAUUAAAACUUUUGCCAUGACGACAGUUUGCCGUG---- .(((((((..........((((((......))))))......((((((((..((((.(.((((....)))).)....)))))))))))))))))))...---- ( -28.00) >DroYak_CAF1 15004 99 - 1 GGGUAAAUAUUUGCUAAGCACAAGAAAUUGCUUGUGAACAUUUGUCGUCGUUUGGUUGAUUUAAGAUUAAAACUUUUGCAAUGACGACAGUGUGCUGUG---- .(((((....))))).((((((((......)))))..(((((.(((((((((.((((.............)))).....)))))))))))))))))...---- ( -24.12) >consensus GGGUAAAUAUUUGCUAAGCACAAGAAAUUGCUUGUGAACAUUUGUCGUCGUUUGGUUGAUUUAAGAUUAAAACUUUUGCAAGGACGACAGUGUGCUGGGUG__ .(((((....)))))...((((((......))))))..(((((((((((((..((((.............))))...))...)))))))).)))......... (-20.22 = -20.42 + 0.20)



| Location | 14,271,414 – 14,271,511 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 82.18 |
| Mean single sequence MFE | -16.54 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.96 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14271414 97 - 22224390 GUUGGCACUCCACACCCAACCC--ACACUCUUCGCCCCCGCUGCCAACGCCCAUUUAAGCUAAAAAGUAAACCAAUUGUUAGUUUUAAUGGAAUUUAUU (((((((...............--.................)))))))..(((((.(((((((.((.........)).))))))).)))))........ ( -17.35) >DroSec_CAF1 12013 93 - 1 GUUGGCACUCCAC--CC-ACCCCCG--CUCUUCGCC-CCGCUGCCAACGACCAUUUAAGCUAAAAAGUAAACCAAUUGUUAGUUUUAAUGGAAUUUAUU (((((((......--..-.......--.........-....)))))))..(((((.(((((((.((.........)).))))))).)))))........ ( -17.69) >DroSim_CAF1 16039 96 - 1 GUUGGCACCCCAC--CC-AUCCCCACACUCCUCGCCCCCGCUGCCAACGACCAUUUAAGCUAAAAAGUAAACCAAUUGUUAGUUUUAAUGGAAUUUAUU (((((((......--..-.......................)))))))..(((((.(((((((.((.........)).))))))).)))))........ ( -17.58) >DroEre_CAF1 14456 77 - 1 GUUGCC--UCCAC--A-----------------CGC-CCCCUGCUCACGCCCAUUUAAGCUAAAAAGUAAACCAAUUGUUAGUUUUAAUGGAAUUUAUU ......--.....--.-----------------.((-.....))......(((((.(((((((.((.........)).))))))).)))))........ ( -9.30) >DroYak_CAF1 15103 95 - 1 GUUGGCACUCCAC--AG-CCCGGCACACUUUUCUGC-UCCCUGCCAACGCCCAUUUAAGCUAAAAAGUAAACCAAUUGUUAGUUUUAAUGGAAUUUAUU (((((((.....(--((-...((....))...))).-....)))))))..(((((.(((((((.((.........)).))))))).)))))........ ( -20.80) >consensus GUUGGCACUCCAC__CC_ACCC_CACACUCUUCGCC_CCGCUGCCAACGCCCAUUUAAGCUAAAAAGUAAACCAAUUGUUAGUUUUAAUGGAAUUUAUU (((((((..................................)))))))..(((((.(((((((.((.........)).))))))).)))))........ (-14.52 = -14.96 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:33 2006