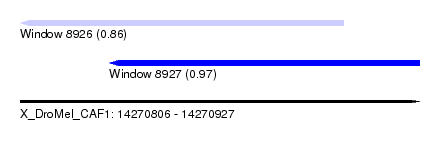
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,270,806 – 14,270,927 |
| Length | 121 |
| Max. P | 0.970940 |

| Location | 14,270,806 – 14,270,904 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.87 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -21.96 |
| Energy contribution | -22.52 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
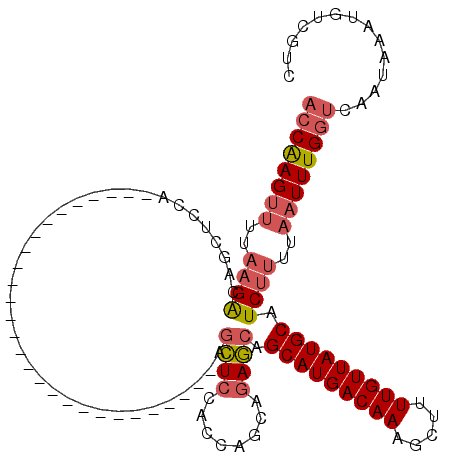
>X_DroMel_CAF1 14270806 98 - 22224390 ---------CAGCUCCACAAGCAGAGCAGCAUGACAAAGCUAUUGUUAUGCAUCUUUUAAUUUGGUCAAUAAAUGUCGUCCCAGGA--AGGACCUUGGGGACGAGGGUG----------- ---------..((((........)))).(((((((((.....)))))))))......................(.((((((((((.--....)))..))))))).)...----------- ( -30.50) >DroPse_CAF1 13734 103 - 1 -----------CAUCCG-----ACAUCAGCAUGACAAAGCUUUUGUUAUGCAUCUUUUAAUUUGGUCAAUAAAUGUCCUUCCCUUCCCUUGCCUUCCCGGA-GGAGGGGGCAGAAACUUU -----------.....(-----((((..(((((((((.....)))))))))(((.........)))......)))))((((((((((....((.....)).-)))))))).))....... ( -27.70) >DroSec_CAF1 11432 97 - 1 ----------AGCUCCACCAGCAGAGCAGCAUGACAAAGCUUUUGUUAUGCAUCUUUUAAUUUGGUCAAUAAAUGUCGUCCCAGGA--AGGACCUUGGGGACGGGGGUG----------- ----------.((((((((((.((((..(((((((((.....)))))))))..))))....)))))..........(((((((((.--....)))..))))))))))).----------- ( -34.00) >DroSim_CAF1 15440 99 - 1 --------CCAGCUCCACCAGCAGAGCAGCAUGACAAAGCUUUUGUUAUGCAUCUUUUAAUUUGGUCAAUAAAUGUCGUCCCAGGA--AGGACCUUGGGGACGGGGAUG----------- --------(((((((........)))).(((((((((.....)))))))))...........)))........(.((((((((((.--....)))..))))))).)...----------- ( -31.50) >DroEre_CAF1 13832 106 - 1 CUCCAUCUCCAGCUCCACCAGCAGAGCAGCAUGACAAAGCUUUUGUUAUGCAUCUUUUAAUUUGGUCAAUAAAUGUCGUCCCAGGA--AGGACCUAGGGGA-GGGGGUG----------- ((((.(((((.((((........)))).(((((((((.....)))))))))............((((......((......))...--..))))..)))))-))))...----------- ( -33.80) >DroYak_CAF1 14479 96 - 1 -----------GCUCCACCAGCAGAGCAGCAUGACAAAGCUUUUGUUAUGCAUCUUUUAAUUUGGUCAAUAAAUGUCGUACGAGGA--AGGACCUUGUGGACGGGGAUG----------- -----------.(((((((((.((((..(((((((((.....)))))))))..))))....)))))........(((.(((((((.--....))))))))))))))...----------- ( -31.40) >consensus __________AGCUCCACCAGCAGAGCAGCAUGACAAAGCUUUUGUUAUGCAUCUUUUAAUUUGGUCAAUAAAUGUCGUCCCAGGA__AGGACCUUGGGGACGGGGGUG___________ ...........((((........)))).(((((((((.....)))))))))((((....(((((.....))))).((((((((((.......)))..)))))))))))............ (-21.96 = -22.52 + 0.56)


| Location | 14,270,833 – 14,270,927 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.09 |
| Mean single sequence MFE | -27.85 |
| Consensus MFE | -21.99 |
| Energy contribution | -22.77 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.82 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14270833 94 - 22224390 ACCAAGUUUUAAGACAGCUCCAG--------------------------CAGCUCCACAAGCAGAGCAGCAUGACAAAGCUAUUGUUAUGCAUCUUUUAAUUUGGUCAAUAAAUGUCGUC ((((((((..((((..((....)--------------------------).((((........)))).(((((((((.....))))))))).))))..)))))))).............. ( -27.90) >DroPse_CAF1 13773 79 - 1 --CGAGG-----GGCAGAAUAU-----------------------------CAUCCG-----ACAUCAGCAUGACAAAGCUUUUGUUAUGCAUCUUUUAAUUUGGUCAAUAAAUGUCCUU --..(((-----((((...(((-----------------------------...(((-----....).(((((((((.....)))))))))............))...)))..))))))) ( -19.40) >DroSec_CAF1 11459 91 - 1 ACCAAGUUUUAAGACAGCUCC-----------------------------AGCUCCACCAGCAGAGCAGCAUGACAAAGCUUUUGUUAUGCAUCUUUUAAUUUGGUCAAUAAAUGUCGUC ((((((((..((((..((((.-----------------------------.(((.....))).)))).(((((((((.....))))))))).))))..)))))))).............. ( -29.40) >DroSim_CAF1 15467 97 - 1 ACCAAGUUUUAAGACAGCUCCAGCU-----------------------CCAGCUCCACCAGCAGAGCAGCAUGACAAAGCUUUUGUUAUGCAUCUUUUAAUUUGGUCAAUAAAUGUCGUC ((((((((..((((........(((-----------------------(..(((.....))).)))).(((((((((.....))))))))).))))..)))))))).............. ( -29.40) >DroEre_CAF1 13858 120 - 1 ACCAAGUUUUAAGACAGCUCCAGCUCCGUUCCAGCUCCAUCUCCAUCUCCAGCUCCACCAGCAGAGCAGCAUGACAAAGCUUUUGUUAUGCAUCUUUUAAUUUGGUCAAUAAAUGUCGUC ((((((((..((((..((((.((((.......))))...............(((.....))).)))).(((((((((.....))))))))).))))..)))))))).............. ( -31.50) >DroYak_CAF1 14506 91 - 1 ACCAAGUUUUAAGACAGUUCCU-----------------------------GCUCCACCAGCAGAGCAGCAUGACAAAGCUUUUGUUAUGCAUCUUUUAAUUUGGUCAAUAAAUGUCGUA ((((((((..((((..((((.(-----------------------------(((.....)))))))).(((((((((.....))))))))).))))..)))))))).............. ( -29.50) >consensus ACCAAGUUUUAAGACAGCUCCA____________________________AGCUCCACCAGCAGAGCAGCAUGACAAAGCUUUUGUUAUGCAUCUUUUAAUUUGGUCAAUAAAUGUCGUC ((((((((..((((.....................................((((........)))).(((((((((.....))))))))).))))..)))))))).............. (-21.99 = -22.77 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:29 2006