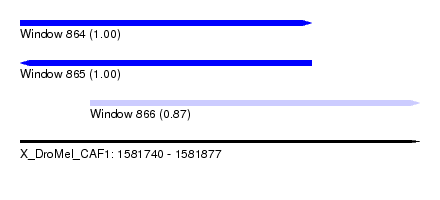
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,581,740 – 1,581,877 |
| Length | 137 |
| Max. P | 0.999544 |

| Location | 1,581,740 – 1,581,840 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 69.21 |
| Mean single sequence MFE | -37.94 |
| Consensus MFE | -14.90 |
| Energy contribution | -17.74 |
| Covariance contribution | 2.84 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.39 |
| SVM decision value | 3.71 |
| SVM RNA-class probability | 0.999544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1581740 100 + 22224390 UGCCG--UAGC----AGCAU----AAUGUCGCAUUCAUUAAGCCAAACAGCAGGUGAUGCACACCUGCGGCUCC--AGCUGC-CUAGCUGCC-UAGCUGCCUGGCUCCCUGCUC .((((--..((----(((.(----((((.......)))))((((.....(((((((.....))))))))))).(--((((..-..)))))..-..))))).))))......... ( -35.90) >DroSec_CAF1 43269 92 + 1 UGCCA--UAGC----AGCAU----AAUGUCGCAUUCAUUAAGCCAAACAGCAGGUGAUGCACACCUGCGGCUCC--AGCUGC-CU---------AGCUGCCUGGCUCCCUGCUU .....--.(((----((..(----((((.......)))))(((((..(((((((((.....)))))(((((...--.)))))-..---------.))))..)))))..))))). ( -34.10) >DroEre_CAF1 46505 102 + 1 UGCCGCACAGC----AGCAU----AAUGUCGCAUUCAUUAAGCCAAACAGCAGGUGAUGCACACCUGCGGCUCC--AGCUGU-GUGGCUGCC-UGGCUGCCUGGCUGCCAGGUC .((((((((((----....(----((((.......)))))((((.....(((((((.....)))))))))))..--.)))))-))))).(((-((((.((...)).))))))). ( -51.80) >DroMoj_CAF1 53938 101 + 1 UGCUCAGUUGUGGCAUGCAUACGGAUUGUCGCAUUCAUUAGGGAAAACAGCAGGUGAUACGCACCUGGUGCGCCGCAGAUGCUCUAGCUGCCAUAGCUGCC------------- ....(((((((((((.((.((.((.((((.((.(((......)))..((.((((((.....)))))).)).)).))))...)).)))))))))))))))..------------- ( -36.80) >DroAna_CAF1 58022 82 + 1 -GCCU--GGGU----G--GU----AAUGUCGCAUUCAUUAAGCCAAACAGCAGGUGAUGCACACCUGCGGCUCC--AGGUGC-UUCUCCUCC-CAGCUA--------------- -((.(--(((.----(--(.----...(..(((((.....((((.....(((((((.....)))))))))))..--.)))))-..).)).))-))))..--------------- ( -31.10) >consensus UGCCG__UAGC____AGCAU____AAUGUCGCAUUCAUUAAGCCAAACAGCAGGUGAUGCACACCUGCGGCUCC__AGCUGC_CUAGCUGCC_UAGCUGCCUGGCU_CC_G_U_ ........................................(((((..(((((((((.....)))))(((((......))))).............))))..)))))........ (-14.90 = -17.74 + 2.84)

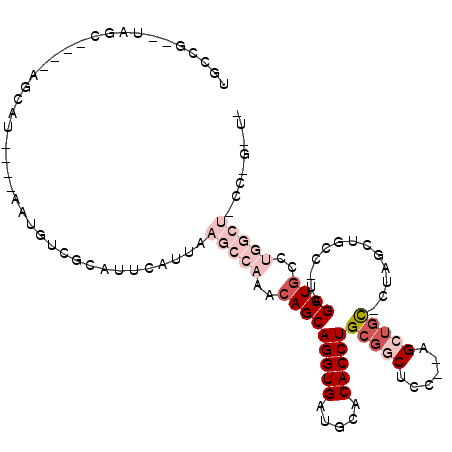
| Location | 1,581,740 – 1,581,840 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 69.21 |
| Mean single sequence MFE | -38.50 |
| Consensus MFE | -18.42 |
| Energy contribution | -19.18 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1581740 100 - 22224390 GAGCAGGGAGCCAGGCAGCUA-GGCAGCUAG-GCAGCU--GGAGCCGCAGGUGUGCAUCACCUGCUGUUUGGCUUAAUGAAUGCGACAUU----AUGCU----GCUA--CGGCA .((((..(((((((((((((.-..(((((..-..))))--).))).(((((((.....)))))))))))))))))((((.......))))----.))))----((..--..)). ( -43.20) >DroSec_CAF1 43269 92 - 1 AAGCAGGGAGCCAGGCAGCU---------AG-GCAGCU--GGAGCCGCAGGUGUGCAUCACCUGCUGUUUGGCUUAAUGAAUGCGACAUU----AUGCU----GCUA--UGGCA .(((((.((((((((((...---------.(-((....--...)))(((((((.....)))))))))))))))))((((.......))))----...))----))).--..... ( -37.90) >DroEre_CAF1 46505 102 - 1 GACCUGGCAGCCAGGCAGCCA-GGCAGCCAC-ACAGCU--GGAGCCGCAGGUGUGCAUCACCUGCUGUUUGGCUUAAUGAAUGCGACAUU----AUGCU----GCUGUGCGGCA (.((((((.((...)).))))-))).(((.(-(((((.--.((((.(((((((.....))))))).))))(((.(((((.......))))----).)))----)))))).))). ( -50.70) >DroMoj_CAF1 53938 101 - 1 -------------GGCAGCUAUGGCAGCUAGAGCAUCUGCGGCGCACCAGGUGCGUAUCACCUGCUGUUUUCCCUAAUGAAUGCGACAAUCCGUAUGCAUGCCACAACUGAGCA -------------((((((.(((((((.....(((((((.(.....)))))))).......)))))))............(((((......))))))).))))........... ( -29.30) >DroAna_CAF1 58022 82 - 1 ---------------UAGCUG-GGAGGAGAA-GCACCU--GGAGCCGCAGGUGUGCAUCACCUGCUGUUUGGCUUAAUGAAUGCGACAUU----AC--C----ACCC--AGGC- ---------------...(((-((.((.((.-(((...--.((((((((((((.....))))))).....)))))......)))....))----.)--)----.)))--))..- ( -31.40) >consensus _A_C_GG_AGCCAGGCAGCUA_GGCAGCUAG_GCAGCU__GGAGCCGCAGGUGUGCAUCACCUGCUGUUUGGCUUAAUGAAUGCGACAUU____AUGCU____GCUA__CGGCA .................(((............(((......((((((((((((.....))))))).....)))))......)))............((.....)).....))). (-18.42 = -19.18 + 0.76)

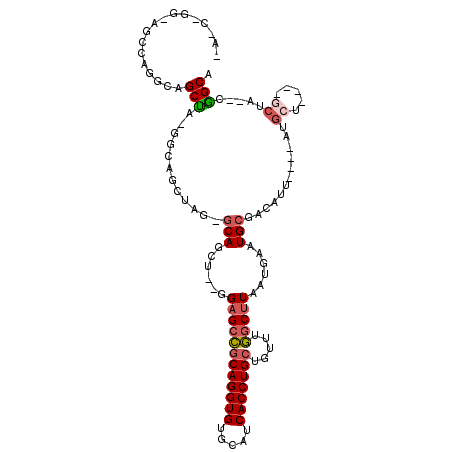
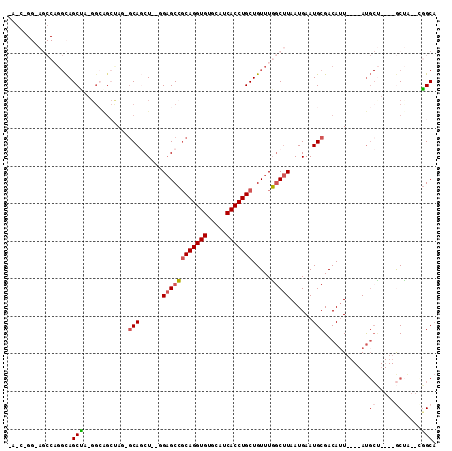
| Location | 1,581,764 – 1,581,877 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.26 |
| Mean single sequence MFE | -37.49 |
| Consensus MFE | -23.33 |
| Energy contribution | -24.70 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.874193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
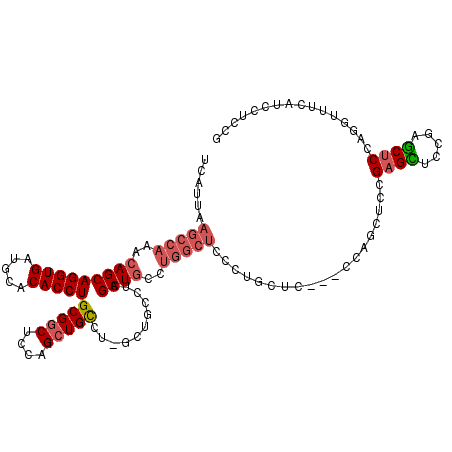
>X_DroMel_CAF1 1581764 113 + 22224390 UCAUUAAGCCAAACAGCAGGUGAUGCACACCUGCGGCUCCAGCUGCCUAGCUGCCUAGCUGCCUGGCUCCCUGCUC---UGAGCUGCGAGCUGCGAGCUCCAGGUUUCGUCCUCCG .....(((((.....(((((((.....)))))))((((((((((((.(((((....)))))...(((((.......---.))))))).))))).)))))...)))))......... ( -42.90) >DroSec_CAF1 43293 105 + 1 UCAUUAAGCCAAACAGCAGGUGAUGCACACCUGCGGCUCCAGCUGCCU--------AGCUGCCUGGCUCCCUGCUU---UGAGCUGCGAGCUUCGAGCUCCAGGUUUCGUCCUCCG ......(((((..(((((((((.....)))))(((((....)))))..--------.))))..))))).((((...---.(((((.((.....)))))))))))............ ( -39.20) >DroEre_CAF1 46531 116 + 1 UCAUUAAGCCAAACAGCAGGUGAUGCACACCUGCGGCUCCAGCUGUGUGGCUGCCUGGCUGCCUGGCUGCCAGGUCCUCCCUGCGCCGAGUUCCGAGCUCUAGGUCUCAUCCUGCG ......(((((.((.(((((((.....)))))))(((....))))).)))))(((((((.((...)).))))))).......((...((((.....)))).(((......))))). ( -45.10) >DroAna_CAF1 58043 92 + 1 UCAUUAAGCCAAACAGCAGGUGAUGCACACCUGCGGCUCCAGGUGCUUCUCCUCCCAGCUA---------------CUCCC----CUGUGUUCUGUACUCUG-----UACUCUGCA ......((((.....(((((((.....))))))))))).(((((((.........(((.((---------------(....----..)))..)))......)-----))).))).. ( -22.76) >consensus UCAUUAAGCCAAACAGCAGGUGAUGCACACCUGCGGCUCCAGCUGCCU_GCUGCCUAGCUGCCUGGCUCCCUGCUC___CCAGCUCCGAGCUCCGAGCUCCAGGUUUCAUCCUCCG ......(((((..(((((((((.....)))))(((((....)))))...........))))..)))))...................((((.....))))................ (-23.33 = -24.70 + 1.37)

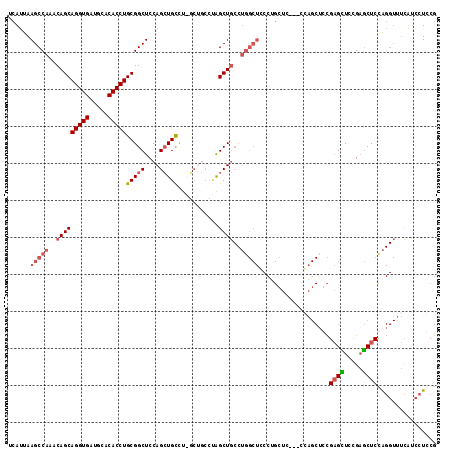
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:30 2006